circstd#
- scipy.stats.circstd(samples, high=6.283185307179586, low=0, axis=None, nan_policy='propagate', *, normalize=False, keepdims=False)[source]#
Compute the circular standard deviation of a sample of angle observations.
Given \(n\) angle observations \(x_1, \cdots, x_n\) measured in radians, their circular standard deviation is defined by ([2], Eq. 2.3.11)
\[\sqrt{ -2 \log \left| \frac{1}{n} \sum_{k=1}^n e^{i x_k} \right| }\]where \(i\) is the imaginary unit and \(|z|\) gives the length of the complex number \(z\). \(|z|\) in the above expression is known as the mean resultant length.
- Parameters:
- samplesarray_like
Input array of angle observations. The value of a full angle is equal to
(high - low).- highfloat, optional
Upper boundary of the principal value of an angle. Default is
2*pi.- lowfloat, optional
Lower boundary of the principal value of an angle. Default is
0.- normalizeboolean, optional
If
False(the default), the return value is computed from the above formula with the input scaled by(2*pi)/(high-low)and the output scaled (back) by(high-low)/(2*pi). IfTrue, the output is not scaled and is returned directly.- axisint or None, default: None
If an int, the axis of the input along which to compute the statistic. The statistic of each axis-slice (e.g. row) of the input will appear in a corresponding element of the output. If
None, the input will be raveled before computing the statistic.- nan_policy{‘propagate’, ‘omit’, ‘raise’}
Defines how to handle input NaNs.
propagate: if a NaN is present in the axis slice (e.g. row) along which the statistic is computed, the corresponding entry of the output will be NaN.omit: NaNs will be omitted when performing the calculation. If insufficient data remains in the axis slice along which the statistic is computed, the corresponding entry of the output will be NaN.raise: if a NaN is present, aValueErrorwill be raised.
- keepdimsbool, default: False
If this is set to True, the axes which are reduced are left in the result as dimensions with size one. With this option, the result will broadcast correctly against the input array.
- Returns:
- circstdfloat
Circular standard deviation, optionally normalized.
If the input array is empty,
np.nanis returned.
Notes
In the limit of small angles, the circular standard deviation is close to the ‘linear’ standard deviation if
normalizeisFalse.Beginning in SciPy 1.9,
np.matrixinputs (not recommended for new code) are converted tonp.ndarraybefore the calculation is performed. In this case, the output will be a scalar ornp.ndarrayof appropriate shape rather than a 2Dnp.matrix. Similarly, while masked elements of masked arrays are ignored, the output will be a scalar ornp.ndarrayrather than a masked array withmask=False.Array API Standard Support
circstdhas experimental support for Python Array API Standard compatible backends in addition to NumPy. Please consider testing these features by setting an environment variableSCIPY_ARRAY_API=1and providing CuPy, PyTorch, JAX, or Dask arrays as array arguments. The following combinations of backend and device (or other capability) are supported.Library
CPU
GPU
NumPy
✅
n/a
CuPy
n/a
✅
PyTorch
✅
✅
JAX
✅
✅
Dask
✅
n/a
See Support for the array API standard for more information.
References
[1]Mardia, K. V. (1972). 2. In Statistics of Directional Data (pp. 18-24). Academic Press. DOI:10.1016/C2013-0-07425-7.
[2]Mardia, K. V. and Jupp, P. E. Directional Statistics. John Wiley & Sons, 1999.
Examples
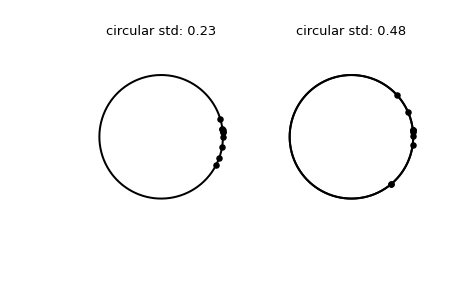
>>> import numpy as np >>> from scipy.stats import circstd >>> import matplotlib.pyplot as plt >>> samples_1 = np.array([0.072, -0.158, 0.077, 0.108, 0.286, ... 0.133, -0.473, -0.001, -0.348, 0.131]) >>> samples_2 = np.array([0.111, -0.879, 0.078, 0.733, 0.421, ... 0.104, -0.136, -0.867, 0.012, 0.105]) >>> circstd_1 = circstd(samples_1) >>> circstd_2 = circstd(samples_2)
Plot the samples.
>>> fig, (left, right) = plt.subplots(ncols=2) >>> for image in (left, right): ... image.plot(np.cos(np.linspace(0, 2*np.pi, 500)), ... np.sin(np.linspace(0, 2*np.pi, 500)), ... c='k') ... image.axis('equal') ... image.axis('off') >>> left.scatter(np.cos(samples_1), np.sin(samples_1), c='k', s=15) >>> left.set_title(f"circular std: {np.round(circstd_1, 2)!r}") >>> right.plot(np.cos(np.linspace(0, 2*np.pi, 500)), ... np.sin(np.linspace(0, 2*np.pi, 500)), ... c='k') >>> right.scatter(np.cos(samples_2), np.sin(samples_2), c='k', s=15) >>> right.set_title(f"circular std: {np.round(circstd_2, 2)!r}") >>> plt.show()