scipy.special.fdtri#
- scipy.special.fdtri(dfn, dfd, p, out=None) = <ufunc 'fdtri'>#
The p-th quantile of the F-distribution.
This function is the inverse of the F-distribution CDF,
fdtr, returning the x such that fdtr(dfn, dfd, x) = p.- Parameters:
- dfnarray_like
First parameter (positive float).
- dfdarray_like
Second parameter (positive float).
- parray_like
Cumulative probability, in [0, 1].
- outndarray, optional
Optional output array for the function values
- Returns:
- xscalar or ndarray
The quantile corresponding to p.
See also
fdtrF distribution cumulative distribution function
fdtrcF distribution survival function
scipy.stats.fF distribution
Notes
The computation is carried out using the relation to the inverse regularized beta function, \(I^{-1}_x(a, b)\). Let \(z = I^{-1}_p(d_d/2, d_n/2).\) Then,
\[x = \frac{d_d (1 - z)}{d_n z}.\]If p is such that \(x < 0.5\), the following relation is used instead for improved stability: let \(z' = I^{-1}_{1 - p}(d_n/2, d_d/2).\) Then,
\[x = \frac{d_d z'}{d_n (1 - z')}.\]Wrapper for the Cephes [1] routine
fdtri.The F distribution is also available as
scipy.stats.f. Callingfdtridirectly can improve performance compared to theppfmethod ofscipy.stats.f(see last example below).References
[1]Cephes Mathematical Functions Library, http://www.netlib.org/cephes/
Examples
fdtrirepresents the inverse of the F distribution CDF which is available asfdtr. Here, we calculate the CDF fordf1=1,df2=2atx=3.fdtrithen returns3given the same values for df1, df2 and the computed CDF value.>>> import numpy as np >>> from scipy.special import fdtri, fdtr >>> df1, df2 = 1, 2 >>> x = 3 >>> cdf_value = fdtr(df1, df2, x) >>> fdtri(df1, df2, cdf_value) 3.000000000000006
Calculate the function at several points by providing a NumPy array for x.
>>> x = np.array([0.1, 0.4, 0.7]) >>> fdtri(1, 2, x) array([0.02020202, 0.38095238, 1.92156863])
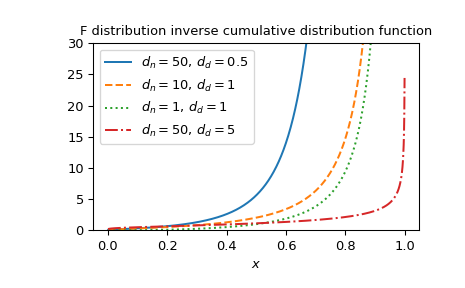
Plot the function for several parameter sets.
>>> import matplotlib.pyplot as plt >>> dfn_parameters = [50, 10, 1, 50] >>> dfd_parameters = [0.5, 1, 1, 5] >>> linestyles = ['solid', 'dashed', 'dotted', 'dashdot'] >>> parameters_list = list(zip(dfn_parameters, dfd_parameters, ... linestyles)) >>> x = np.linspace(0, 1, 1000) >>> fig, ax = plt.subplots() >>> for parameter_set in parameters_list: ... dfn, dfd, style = parameter_set ... fdtri_vals = fdtri(dfn, dfd, x) ... ax.plot(x, fdtri_vals, label=rf"$d_n={dfn},\, d_d={dfd}$", ... ls=style) >>> ax.legend() >>> ax.set_xlabel("$x$") >>> title = "F distribution inverse cumulative distribution function" >>> ax.set_title(title) >>> ax.set_ylim(0, 30) >>> plt.show()
The F distribution is also available as
scipy.stats.f. Usingfdtridirectly can be much faster than calling theppfmethod ofscipy.stats.f, especially for small arrays or individual values. To get the same results one must use the following parametrization:stats.f(dfn, dfd).ppf(x)=fdtri(dfn, dfd, x).>>> from scipy.stats import f >>> dfn, dfd = 1, 2 >>> x = 0.7 >>> fdtri_res = fdtri(dfn, dfd, x) # this will often be faster than below >>> f_dist_res = f(dfn, dfd).ppf(x) >>> f_dist_res == fdtri_res # test that results are equal True