power#
- scipy.stats.power(test, rvs, n_observations, *, significance=0.01, vectorized=None, n_resamples=10000, batch=None, kwargs=None)[source]#
Simulate the power of a hypothesis test under an alternative hypothesis.
- Parameters:
- testcallable
Hypothesis test for which the power is to be simulated.
testmust be a callable that accepts a sample (e.g.test(sample)) orlen(rvs)separate samples (e.g.test(samples1, sample2)if rvs contains two callables and n_observations contains two values) and returns the p-value of the test. If vectorized is set toTrue,testmust also accept a keyword argument axis and be vectorized to perform the test along the provided axis of the samples. Any callable fromscipy.statswith an axis argument that returns an object with a pvalue attribute is also acceptable.- rvscallable or tuple of callables
A callable or sequence of callables that generate(s) random variates under the alternative hypothesis. Each element of rvs must accept keyword argument
size(e.g.rvs(size=(m, n))) and return an N-d array of that shape. If rvs is a sequence, the number of callables in rvs must match the number of elements of n_observations, i.e.len(rvs) == len(n_observations). If rvs is a single callable, n_observations is treated as a single element.- n_observationstuple of ints or tuple of integer arrays
If a sequence of ints, each is the sizes of a sample to be passed to
test. If a sequence of integer arrays, the power is simulated for each set of corresponding sample sizes. See Examples.- significancefloat or array_like of floats, default: 0.01
The threshold for significance; i.e., the p-value below which the hypothesis test results will be considered as evidence against the null hypothesis. Equivalently, the acceptable rate of Type I error under the null hypothesis. If an array, the power is simulated for each significance threshold.
- kwargsdict, optional
Keyword arguments to be passed to rvs and/or
testcallables. Introspection is used to determine which keyword arguments may be passed to each callable. The value corresponding with each keyword must be an array. Arrays must be broadcastable with one another and with each array in n_observations. The power is simulated for each set of corresponding sample sizes and arguments. See Examples.- vectorizedbool, optional
If vectorized is set to
False,testwill not be passed keyword argument axis and is expected to perform the test only for 1D samples. IfTrue,testwill be passed keyword argument axis and is expected to perform the test along axis when passed N-D sample arrays. IfNone(default), vectorized will be setTrueifaxisis a parameter oftest. Use of a vectorized test typically reduces computation time.- n_resamplesint, default: 10000
Number of samples drawn from each of the callables of rvs. Equivalently, the number tests performed under the alternative hypothesis to approximate the power.
- batchint, optional
The number of samples to process in each call to
test. Memory usage is proportional to the product of batch and the largest sample size. Default isNone, in which case batch equals n_resamples.
- Returns:
- resPowerResult
An object with attributes:
- powerfloat or ndarray
The estimated power against the alternative.
- pvaluesndarray
The p-values observed under the alternative hypothesis.
Notes
The power is simulated as follows:
Draw many random samples (or sets of samples), each of the size(s) specified by n_observations, under the alternative specified by rvs.
For each sample (or set of samples), compute the p-value according to
test. These p-values are recorded in thepvaluesattribute of the result object.Compute the proportion of p-values that are less than the significance level. This is the power recorded in the
powerattribute of the result object.
Suppose that significance is an array with shape
shape1, the elements of kwargs and n_observations are mutually broadcastable to shapeshape2, andtestreturns an array of p-values of shapeshape3. Then the result objectpowerattribute will be of shapeshape1 + shape2 + shape3, and thepvaluesattribute will be of shapeshape2 + shape3 + (n_resamples,).Array API Standard Support
powerhas experimental support for Python Array API Standard compatible backends in addition to NumPy. Please consider testing these features by setting an environment variableSCIPY_ARRAY_API=1and providing CuPy, PyTorch, JAX, or Dask arrays as array arguments. The following combinations of backend and device (or other capability) are supported.Library
CPU
GPU
NumPy
✅
n/a
CuPy
n/a
✅
PyTorch
✅
✅
JAX
⚠️ no JIT
⚠️ no JIT
Dask
⚠️ computes graph
n/a
See Support for the array API standard for more information.
Examples
Suppose we wish to simulate the power of the independent sample t-test under the following conditions:
The first sample has 10 observations drawn from a normal distribution with mean 0.
The second sample has 12 observations drawn from a normal distribution with mean 1.0.
The threshold on p-values for significance is 0.05.
>>> import numpy as np >>> from scipy import stats >>> rng = np.random.default_rng() >>> >>> test = stats.ttest_ind >>> n_observations = (10, 12) >>> rvs1 = rng.normal >>> rvs2 = lambda size: rng.normal(loc=1, size=size) >>> rvs = (rvs1, rvs2) >>> res = stats.power(test, rvs, n_observations, significance=0.05) >>> res.power 0.6116
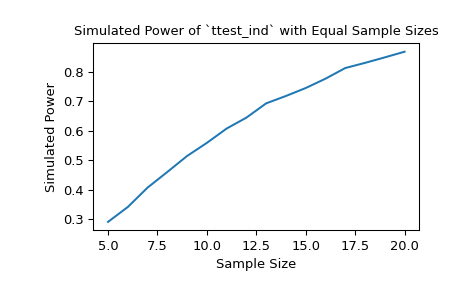
With samples of size 10 and 12, respectively, the power of the t-test with a significance threshold of 0.05 is approximately 60% under the chosen alternative. We can investigate the effect of sample size on the power by passing sample size arrays.
>>> import matplotlib.pyplot as plt >>> nobs_x = np.arange(5, 21) >>> nobs_y = nobs_x >>> n_observations = (nobs_x, nobs_y) >>> res = stats.power(test, rvs, n_observations, significance=0.05) >>> ax = plt.subplot() >>> ax.plot(nobs_x, res.power) >>> ax.set_xlabel('Sample Size') >>> ax.set_ylabel('Simulated Power') >>> ax.set_title('Simulated Power of `ttest_ind` with Equal Sample Sizes') >>> plt.show()
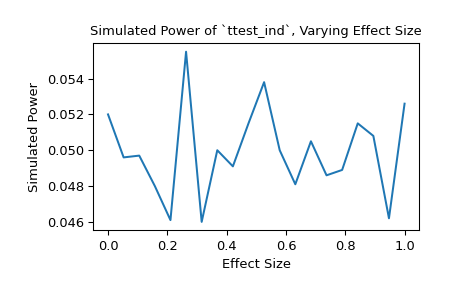
Alternatively, we can investigate the impact that effect size has on the power. In this case, the effect size is the location of the distribution underlying the second sample.
>>> n_observations = (10, 12) >>> loc = np.linspace(0, 1, 20) >>> rvs2 = lambda size, loc: rng.normal(loc=loc, size=size) >>> rvs = (rvs1, rvs2) >>> res = stats.power(test, rvs, n_observations, significance=0.05, ... kwargs={'loc': loc}) >>> ax = plt.subplot() >>> ax.plot(loc, res.power) >>> ax.set_xlabel('Effect Size') >>> ax.set_ylabel('Simulated Power') >>> ax.set_title('Simulated Power of `ttest_ind`, Varying Effect Size') >>> plt.show()
We can also use
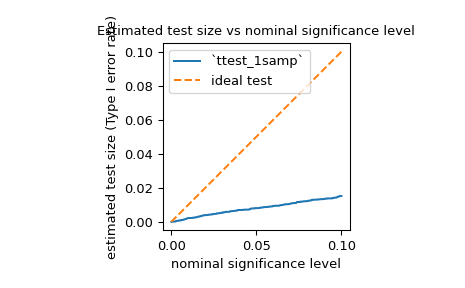
powerto estimate the Type I error rate (also referred to by the ambiguous term “size”) of a test and assess whether it matches the nominal level. For example, the null hypothesis ofjarque_berais that the sample was drawn from a distribution with the same skewness and kurtosis as the normal distribution. To estimate the Type I error rate, we can consider the null hypothesis to be a true alternative hypothesis and calculate the power.>>> test = stats.jarque_bera >>> n_observations = 10 >>> rvs = rng.normal >>> significance = np.linspace(0.0001, 0.1, 1000) >>> res = stats.power(test, rvs, n_observations, significance=significance) >>> size = res.power
As shown below, the Type I error rate of the test is far below the nominal level for such a small sample, as mentioned in its documentation.
>>> ax = plt.subplot() >>> ax.plot(significance, size) >>> ax.plot([0, 0.1], [0, 0.1], '--') >>> ax.set_xlabel('nominal significance level') >>> ax.set_ylabel('estimated test size (Type I error rate)') >>> ax.set_title('Estimated test size vs nominal significance level') >>> ax.set_aspect('equal', 'box') >>> ax.legend(('`ttest_1samp`', 'ideal test')) >>> plt.show()
As one might expect from such a conservative test, the power is quite low with respect to some alternatives. For example, the power of the test under the alternative that the sample was drawn from the Laplace distribution may not be much greater than the Type I error rate.
>>> rvs = rng.laplace >>> significance = np.linspace(0.0001, 0.1, 1000) >>> res = stats.power(test, rvs, n_observations, significance=0.05) >>> print(res.power) 0.0587
This is not a mistake in SciPy’s implementation; it is simply due to the fact that the null distribution of the test statistic is derived under the assumption that the sample size is large (i.e. approaches infinity), and this asymptotic approximation is not accurate for small samples. In such cases, resampling and Monte Carlo methods (e.g.
permutation_test,goodness_of_fit,monte_carlo_test) may be more appropriate.