scipy.stats.truncpareto#
- scipy.stats.truncpareto = <scipy.stats._continuous_distns.truncpareto_gen object>[source]#
An upper truncated Pareto continuous random variable.
As an instance of the
rv_continuousclass,truncparetoobject inherits from it a collection of generic methods (see below for the full list), and completes them with details specific for this particular distribution.Methods
rvs(b, c, loc=0, scale=1, size=1, random_state=None)
Random variates.
pdf(x, b, c, loc=0, scale=1)
Probability density function.
logpdf(x, b, c, loc=0, scale=1)
Log of the probability density function.
cdf(x, b, c, loc=0, scale=1)
Cumulative distribution function.
logcdf(x, b, c, loc=0, scale=1)
Log of the cumulative distribution function.
sf(x, b, c, loc=0, scale=1)
Survival function (also defined as
1 - cdf, but sf is sometimes more accurate).logsf(x, b, c, loc=0, scale=1)
Log of the survival function.
ppf(q, b, c, loc=0, scale=1)
Percent point function (inverse of
cdf— percentiles).isf(q, b, c, loc=0, scale=1)
Inverse survival function (inverse of
sf).moment(order, b, c, loc=0, scale=1)
Non-central moment of the specified order.
stats(b, c, loc=0, scale=1, moments=’mv’)
Mean(‘m’), variance(‘v’), skew(‘s’), and/or kurtosis(‘k’).
entropy(b, c, loc=0, scale=1)
(Differential) entropy of the RV.
fit(data)
Parameter estimates for generic data. See scipy.stats.rv_continuous.fit for detailed documentation of the keyword arguments.
expect(func, args=(b, c), loc=0, scale=1, lb=None, ub=None, conditional=False, **kwds)
Expected value of a function (of one argument) with respect to the distribution.
median(b, c, loc=0, scale=1)
Median of the distribution.
mean(b, c, loc=0, scale=1)
Mean of the distribution.
var(b, c, loc=0, scale=1)
Variance of the distribution.
std(b, c, loc=0, scale=1)
Standard deviation of the distribution.
interval(confidence, b, c, loc=0, scale=1)
Confidence interval with equal areas around the median.
See also
paretoPareto distribution
Notes
The probability density function for
truncparetois:\[f(x, b, c) = \frac{b}{1 - c^{-b}} \frac{1}{x^{b+1}}\]for \(b > 0\), \(c > 1\) and \(1 \le x \le c\).
truncparetotakes b and c as shape parameters for \(b\) and \(c\).Notice that the upper truncation value \(c\) is defined in standardized form so that random values of an unscaled, unshifted variable are within the range
[1, c]. Ifu_ris the upper bound to a scaled and/or shifted variable, thenc = (u_r - loc) / scale. In other words, the support of the distribution becomes(scale + loc) <= x <= (c*scale + loc)when scale and/or loc are provided.The probability density above is defined in the “standardized” form. To shift and/or scale the distribution use the
locandscaleparameters. Specifically,truncpareto.pdf(x, b, c, loc, scale)is identically equivalent totruncpareto.pdf(y, b, c) / scalewithy = (x - loc) / scale. Note that shifting the location of a distribution does not make it a “noncentral” distribution; noncentral generalizations of some distributions are available in separate classes.References
[1]Burroughs, S. M., and Tebbens S. F. “Upper-truncated power laws in natural systems.” Pure and Applied Geophysics 158.4 (2001): 741-757.
Examples
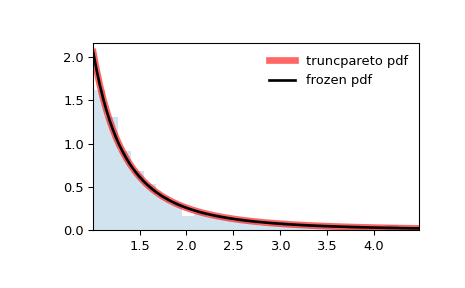
>>> import numpy as np >>> from scipy.stats import truncpareto >>> import matplotlib.pyplot as plt >>> fig, ax = plt.subplots(1, 1)
Calculate the first four moments:
>>> b, c = 2, 5 >>> mean, var, skew, kurt = truncpareto.stats(b, c, moments='mvsk')
Display the probability density function (
pdf):>>> x = np.linspace(truncpareto.ppf(0.01, b, c), ... truncpareto.ppf(0.99, b, c), 100) >>> ax.plot(x, truncpareto.pdf(x, b, c), ... 'r-', lw=5, alpha=0.6, label='truncpareto pdf')
Alternatively, the distribution object can be called (as a function) to fix the shape, location and scale parameters. This returns a “frozen” RV object holding the given parameters fixed.
Freeze the distribution and display the frozen
pdf:>>> rv = truncpareto(b, c) >>> ax.plot(x, rv.pdf(x), 'k-', lw=2, label='frozen pdf')
Check accuracy of
cdfandppf:>>> vals = truncpareto.ppf([0.001, 0.5, 0.999], b, c) >>> np.allclose([0.001, 0.5, 0.999], truncpareto.cdf(vals, b, c)) True
Generate random numbers:
>>> r = truncpareto.rvs(b, c, size=1000)
And compare the histogram:
>>> ax.hist(r, density=True, bins='auto', histtype='stepfilled', alpha=0.2) >>> ax.set_xlim([x[0], x[-1]]) >>> ax.legend(loc='best', frameon=False) >>> plt.show()