MultinomialQMC#
- class scipy.stats.qmc.MultinomialQMC(pvals, n_trials, *, engine=None, seed=None)[source]#
QMC sampling from a multinomial distribution.
- Parameters:
- pvalsarray_like (k,)
Vector of probabilities of size
k, wherekis the number of categories. Elements must be non-negative and sum to 1.- n_trialsint
Number of trials.
- engineQMCEngine, optional
Quasi-Monte Carlo engine sampler. If None,
Sobolis used.- seed{None, int,
numpy.random.Generator}, optional Used only if engine is None. If seed is an int or None, a new
numpy.random.Generatoris created usingnp.random.default_rng(seed). If seed is already aGeneratorinstance, then the provided instance is used.
Methods
random([n])Draw n QMC samples from the multinomial distribution.
Examples
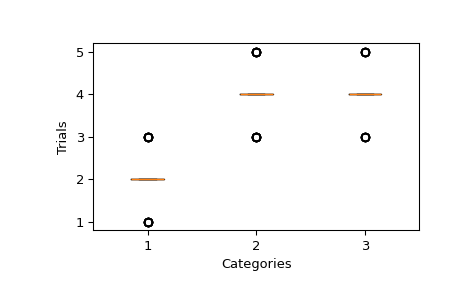
Let’s define 3 categories and for a given sample, the sum of the trials of each category is 8. The number of trials per category is determined by the pvals associated to each category. Then, we sample this distribution 64 times.
>>> import matplotlib.pyplot as plt >>> from scipy.stats import qmc >>> dist = qmc.MultinomialQMC( ... pvals=[0.2, 0.4, 0.4], n_trials=10, engine=qmc.Halton(d=1) ... ) >>> sample = dist.random(64)
We can plot the sample and verify that the median of number of trials for each category is following the pvals. That would be
pvals * n_trials = [2, 4, 4].>>> fig, ax = plt.subplots() >>> ax.yaxis.get_major_locator().set_params(integer=True) >>> _ = ax.boxplot(sample) >>> ax.set(xlabel="Categories", ylabel="Trials") >>> plt.show()