scipy.signal.stft#
- scipy.signal.stft(x, fs=1.0, window='hann', nperseg=256, noverlap=None, nfft=None, detrend=False, return_onesided=True, boundary='zeros', padded=True, axis=-1, scaling='spectrum')[source]#
Compute the Short Time Fourier Transform (STFT).
STFTs can be used as a way of quantifying the change of a nonstationary signal’s frequency and phase content over time.
- Parameters:
- xarray_like
Time series of measurement values
- fsfloat, optional
Sampling frequency of the x time series. Defaults to 1.0.
- windowstr or tuple or array_like, optional
Desired window to use. If window is a string or tuple, it is passed to
get_windowto generate the window values, which are DFT-even by default. Seeget_windowfor a list of windows and required parameters. If window is array_like it will be used directly as the window and its length must be nperseg. Defaults to a Hann window.- npersegint, optional
Length of each segment. Defaults to 256.
- noverlapint, optional
Number of points to overlap between segments. If None,
noverlap = nperseg // 2. Defaults to None. When specified, the COLA constraint must be met (see Notes below).- nfftint, optional
Length of the FFT used, if a zero padded FFT is desired. If None, the FFT length is nperseg. Defaults to None.
- detrendstr or function or False, optional
Specifies how to detrend each segment. If
detrendis a string, it is passed as the type argument to thedetrendfunction. If it is a function, it takes a segment and returns a detrended segment. Ifdetrendis False, no detrending is done. Defaults to False.- return_onesidedbool, optional
If True, return a one-sided spectrum for real data. If False return a two-sided spectrum. Defaults to True, but for complex data, a two-sided spectrum is always returned.
- boundarystr or None, optional
Specifies whether the input signal is extended at both ends, and how to generate the new values, in order to center the first windowed segment on the first input point. This has the benefit of enabling reconstruction of the first input point when the employed window function starts at zero. Valid options are
['even', 'odd', 'constant', 'zeros', None]. Defaults to ‘zeros’, for zero padding extension. I.e.[1, 2, 3, 4]is extended to[0, 1, 2, 3, 4, 0]fornperseg=3.- paddedbool, optional
Specifies whether the input signal is zero-padded at the end to make the signal fit exactly into an integer number of window segments, so that all of the signal is included in the output. Defaults to True. Padding occurs after boundary extension, if boundary is not None, and padded is True, as is the default.
- axisint, optional
Axis along which the STFT is computed; the default is over the last axis (i.e.
axis=-1).- scaling: {‘spectrum’, ‘psd’}
The default ‘spectrum’ scaling allows each frequency line of Zxx to be interpreted as a magnitude spectrum. The ‘psd’ option scales each line to a power spectral density - it allows to calculate the signal’s energy by numerically integrating over
abs(Zxx)**2.New in version 1.9.0.
- Returns:
- fndarray
Array of sample frequencies.
- tndarray
Array of segment times.
- Zxxndarray
STFT of x. By default, the last axis of Zxx corresponds to the segment times.
See also
istftInverse Short Time Fourier Transform
check_COLACheck whether the Constant OverLap Add (COLA) constraint is met
check_NOLACheck whether the Nonzero Overlap Add (NOLA) constraint is met
welchPower spectral density by Welch’s method.
spectrogramSpectrogram by Welch’s method.
csdCross spectral density by Welch’s method.
lombscargleLomb-Scargle periodogram for unevenly sampled data
Notes
In order to enable inversion of an STFT via the inverse STFT in
istft, the signal windowing must obey the constraint of “Nonzero OverLap Add” (NOLA), and the input signal must have complete windowing coverage (i.e.(x.shape[axis] - nperseg) % (nperseg-noverlap) == 0). The padded argument may be used to accomplish this.Given a time-domain signal \(x[n]\), a window \(w[n]\), and a hop size \(H\) = nperseg - noverlap, the windowed frame at time index \(t\) is given by
\[x_{t}[n]=x[n]w[n-tH]\]The overlap-add (OLA) reconstruction equation is given by
\[x[n]=\frac{\sum_{t}x_{t}[n]w[n-tH]}{\sum_{t}w^{2}[n-tH]}\]The NOLA constraint ensures that every normalization term that appears in the denomimator of the OLA reconstruction equation is nonzero. Whether a choice of window, nperseg, and noverlap satisfy this constraint can be tested with
check_NOLA.New in version 0.19.0.
References
[1]Oppenheim, Alan V., Ronald W. Schafer, John R. Buck “Discrete-Time Signal Processing”, Prentice Hall, 1999.
[2]Daniel W. Griffin, Jae S. Lim “Signal Estimation from Modified Short-Time Fourier Transform”, IEEE 1984, 10.1109/TASSP.1984.1164317
Examples
>>> import numpy as np >>> from scipy import signal >>> import matplotlib.pyplot as plt >>> rng = np.random.default_rng()
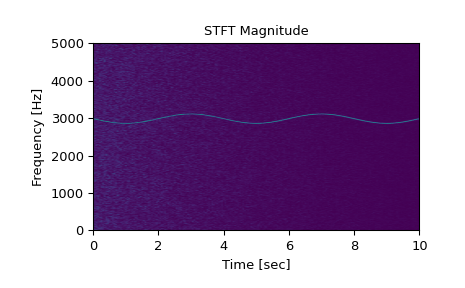
Generate a test signal, a 2 Vrms sine wave whose frequency is slowly modulated around 3kHz, corrupted by white noise of exponentially decreasing magnitude sampled at 10 kHz.
>>> fs = 10e3 >>> N = 1e5 >>> amp = 2 * np.sqrt(2) >>> noise_power = 0.01 * fs / 2 >>> time = np.arange(N) / float(fs) >>> mod = 500*np.cos(2*np.pi*0.25*time) >>> carrier = amp * np.sin(2*np.pi*3e3*time + mod) >>> noise = rng.normal(scale=np.sqrt(noise_power), ... size=time.shape) >>> noise *= np.exp(-time/5) >>> x = carrier + noise
Compute and plot the STFT’s magnitude.
>>> f, t, Zxx = signal.stft(x, fs, nperseg=1000) >>> plt.pcolormesh(t, f, np.abs(Zxx), vmin=0, vmax=amp, shading='gouraud') >>> plt.title('STFT Magnitude') >>> plt.ylabel('Frequency [Hz]') >>> plt.xlabel('Time [sec]') >>> plt.show()
Compare the energy of the signal x with the energy of its STFT:
>>> E_x = sum(x**2) / fs # Energy of x >>> # Calculate a two-sided STFT with PSD scaling: >>> f, t, Zxx = signal.stft(x, fs, nperseg=1000, return_onesided=False, ... scaling='psd') >>> # Integrate numerically over abs(Zxx)**2: >>> df, dt = f[1] - f[0], t[1] - t[0] >>> E_Zxx = sum(np.sum(Zxx.real**2 + Zxx.imag**2, axis=0) * df) * dt >>> # The energy is the same, but the numerical errors are quite large: >>> np.isclose(E_x, E_Zxx, rtol=1e-2) True