Smoothing splines#
For the interpolation problem, the task is to construct a curve which passes
through a given set of data points. This may be not appropriate if the data is
noisy: we then want to construct a smooth curve, g(x), which approximates
the input data without passing through each point exactly.
To this end, scipy.interpolate allows constructing smoothing splines, based
on the Fortran library FITPACK by P. Dierckx.
Specifically, given the data arrays x and y and the array of
non-negative weights, w, we look for a spline function g(x) which
satisfies
where s is the input parameter which controls the interplay between the
smoothness of the resulting curve g(x) and the quality of the approximation
of the data (i.e., the differences between \(g(x_j)\) and \(y_j\)).
Note that the limit s = 0 corresponds to the interpolation problem where
\(g(x_j) = y_j\). Increasing s leads to smoother fits, and in the limit
of a very large s, \(g(x)\) degenerates into a single best-fit polynomial.
Finding a good value of the s parameter is a trial-and-error process. If
the weights correspond to the inverse of standard deviations of the input data,
the “good” value of s is expected to be somewhere between \(m - \sqrt{2m}\)
and \(m + \sqrt{2m}\), where \(m\) is the number of data points.
If all weights equal unity, a reasonable choice might be around \(s \sim m\,\sigma^2\),
where \(\sigma\) is an estimate for the standard deviation of the data.
Internally, the FITPACK library works by adding internal knots to the spline
fit g(x), so that the resulting knots do not necessarily coincide with the input data.
Spline smoothing in 1-D#
scipy.interpolate provides two interfaces for the FITPACK library, a functional
interface and an object-oriented interface. While equivalent, these interfaces
have different defaults. Below we discuss them in turn, starting from the
functional interface — which we recommend for use in new code.
Procedural (splrep)#
Spline interpolation requires two essential steps: (1) a spline
representation of the curve is computed, and (2) the spline is
evaluated at the desired points. In order to find the spline
representation, there are two different ways to represent a curve and
obtain (smoothing) spline coefficients: directly and parametrically.
The direct method finds the spline representation of a curve in a 2-D
plane using the function splrep. The
first two arguments are the only ones required, and these provide the
\(x\) and \(y\) components of the curve. The normal output is
a 3-tuple, \(\left(t,c,k\right)\) , containing the knot-points,
\(t\) , the coefficients \(c\) and the order \(k\) of the
spline. The default spline order is cubic, but this can be changed
with the input keyword, k.
The knot array defines the interpolation interval to be t[k:-k], so that
the first \(k+1\) and last \(k+1\) entries of the t array define
boundary knots. The coefficients are a 1D array of length at least
len(t) - k - 1. Some routines pad this array to have len(c) == len(t)—
these additional coefficients are ignored for the spline evaluation.
The tck-tuple format is compatible with
interpolating b-splines: the output of
splrep can be wrapped into a BSpline object, e.g. BSpline(*tck), and
the evaluation/integration/root-finding routines described below
can use tck-tuples and BSpline objects interchangeably.
For curves in N-D space the function
splprep allows defining the curve
parametrically. For this function only 1 input argument is
required. This input is a list of \(N\) arrays representing the
curve in N-D space. The length of each array is the
number of curve points, and each array provides one component of the
N-D data point. The parameter variable is given
with the keyword argument, u, which defaults to an equally-spaced
monotonic sequence between \(0\) and \(1\) (i.e., the uniform
parametrization).
The output consists of two objects: a 3-tuple, \(\left(t,c,k\right)\) , containing the spline representation and the parameter variable \(u.\)
The coefficients are a list of \(N\) arrays, where each array corresponds to
a dimension of the input data. Note that the knots, t correspond to the
parametrization of the curve u.
The keyword argument, s , is used to specify the amount of smoothing
to perform during the spline fit. The default value of \(s\) is
\(s=m-\sqrt{2m}\) where \(m\) is the number of data points
being fit. Therefore, if no smoothing is desired a value of
\(\mathbf{s}=0\) should be passed to the routines.
Once the spline representation of the data has been determined, it can be
evaluated either using the splev function or by wrapping
the tck tuple into a BSpline object, as demonstrated below.
We start by illustrating the effect of the s parameter on smoothing some
synthetic noisy data
>>> import numpy as np
>>> from scipy.interpolate import splrep, BSpline
Generate some noisy data
>>> x = np.arange(0, 2*np.pi+np.pi/4, 2*np.pi/16)
>>> rng = np.random.default_rng()
>>> y = np.sin(x) + 0.4*rng.standard_normal(size=len(x))
Construct two splines with different values of s.
>>> tck = splrep(x, y, s=0)
>>> tck_s = splrep(x, y, s=len(x))
And plot them
>>> import matplotlib.pyplot as plt
>>> xnew = np.arange(0, 9/4, 1/50) * np.pi
>>> plt.plot(xnew, np.sin(xnew), '-.', label='sin(x)')
>>> plt.plot(xnew, BSpline(*tck)(xnew), '-', label='s=0')
>>> plt.plot(xnew, BSpline(*tck_s)(xnew), '-', label=f's={len(x)}')
>>> plt.plot(x, y, 'o')
>>> plt.legend()
>>> plt.show()
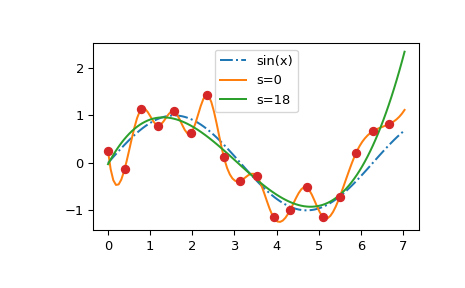
We see that the s=0 curve follows the (random) fluctuations of the data points,
while the s > 0 curve is close to the underlying sine function.
Also note that the extrapolated values vary wildly depending on the value of s.
The default value of s depends on whether the weights are supplied or not,
and also differs for splrep and splprep. Therefore, we recommend always
providing the value of s explicitly.
Manipulating spline objects: procedural (splXXX)#
Once the spline representation of the data has been determined,
functions are available for evaluating the spline
(splev) and its derivatives
(splev, spalde) at any point
and the integral of the spline between any two points (
splint). In addition, for cubic splines ( \(k=3\)
) with 8 or more knots, the roots of the spline can be estimated (
sproot). These functions are demonstrated in the
example that follows.
>>> import numpy as np
>>> import matplotlib.pyplot as plt
>>> from scipy import interpolate
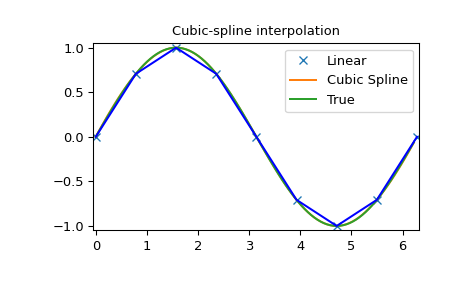
Cubic spline
>>> x = np.arange(0, 2*np.pi+np.pi/4, 2*np.pi/8)
>>> y = np.sin(x)
>>> tck = interpolate.splrep(x, y, s=0)
>>> xnew = np.arange(0, 2*np.pi, np.pi/50)
>>> ynew = interpolate.splev(xnew, tck, der=0)
Note that the last line is equivalent to BSpline(*tck)(xnew).
>>> plt.figure()
>>> plt.plot(x, y, 'x', xnew, ynew, xnew, np.sin(xnew), x, y, 'b')
>>> plt.legend(['Linear', 'Cubic Spline', 'True'])
>>> plt.axis([-0.05, 6.33, -1.05, 1.05])
>>> plt.title('Cubic-spline interpolation')
>>> plt.show()
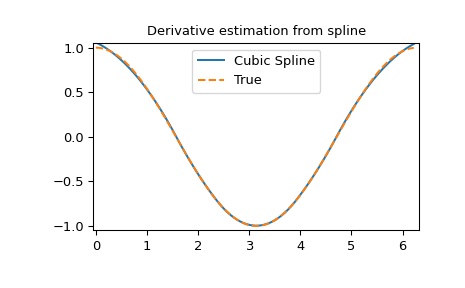
Derivative of spline
>>> yder = interpolate.splev(xnew, tck, der=1) # or BSpline(*tck)(xnew, 1)
>>> plt.figure()
>>> plt.plot(xnew, yder, xnew, np.cos(xnew),'--')
>>> plt.legend(['Cubic Spline', 'True'])
>>> plt.axis([-0.05, 6.33, -1.05, 1.05])
>>> plt.title('Derivative estimation from spline')
>>> plt.show()
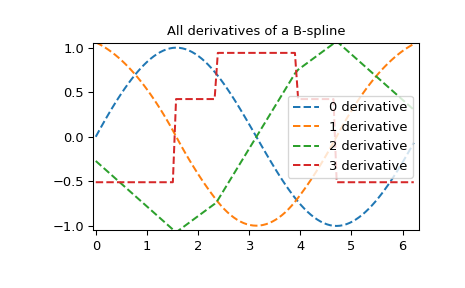
All derivatives of spline
>>> yders = interpolate.spalde(xnew, tck)
>>> plt.figure()
>>> for i in range(len(yders[0])):
... plt.plot(xnew, [d[i] for d in yders], '--', label=f"{i} derivative")
>>> plt.legend()
>>> plt.axis([-0.05, 6.33, -1.05, 1.05])
>>> plt.title('All derivatives of a B-spline')
>>> plt.show()
Integral of spline
>>> def integ(x, tck, constant=-1):
... x = np.atleast_1d(x)
... out = np.zeros(x.shape, dtype=x.dtype)
... for n in range(len(out)):
... out[n] = interpolate.splint(0, x[n], tck)
... out += constant
... return out
>>> yint = integ(xnew, tck)
>>> plt.figure()
>>> plt.plot(xnew, yint, xnew, -np.cos(xnew), '--')
>>> plt.legend(['Cubic Spline', 'True'])
>>> plt.axis([-0.05, 6.33, -1.05, 1.05])
>>> plt.title('Integral estimation from spline')
>>> plt.show()
Roots of spline
>>> interpolate.sproot(tck)
array([3.1416]) # may vary
Notice that sproot may fail to find an obvious solution at the edge of the
approximation interval, \(x = 0\). If we define the spline on a slightly
larger interval, we recover both roots \(x = 0\) and \(x = \pi\):
>>> x = np.linspace(-np.pi/4, np.pi + np.pi/4, 51)
>>> y = np.sin(x)
>>> tck = interpolate.splrep(x, y, s=0)
>>> interpolate.sproot(tck)
array([0., 3.1416])
Parametric spline
>>> t = np.arange(0, 1.1, .1)
>>> x = np.sin(2*np.pi*t)
>>> y = np.cos(2*np.pi*t)
>>> tck, u = interpolate.splprep([x, y], s=0)
>>> unew = np.arange(0, 1.01, 0.01)
>>> out = interpolate.splev(unew, tck)
>>> plt.figure()
>>> plt.plot(x, y, 'x', out[0], out[1], np.sin(2*np.pi*unew), np.cos(2*np.pi*unew), x, y, 'b')
>>> plt.legend(['Linear', 'Cubic Spline', 'True'])
>>> plt.axis([-1.05, 1.05, -1.05, 1.05])
>>> plt.title('Spline of parametrically-defined curve')
>>> plt.show()
Note that in the last example, splprep returns the spline coefficients as a
list of arrays, where each array corresponds to a dimension of the input data.
Thus to wrap its output to a BSpline, we need to transpose the coefficients
(or use BSpline(..., axis=1)):
>>> tt, cc, k = tck
>>> cc = np.array(cc)
>>> bspl = BSpline(tt, cc.T, k) # note the transpose
>>> xy = bspl(u)
>>> xx, yy = xy.T # transpose to unpack into a pair of arrays
>>> np.allclose(x, xx)
True
>>> np.allclose(y, yy)
True
Object-oriented (UnivariateSpline)#
The spline-fitting capabilities described above are also available via
an objected-oriented interface. The 1-D splines are
objects of the UnivariateSpline class, and are created with the
\(x\) and \(y\) components of the curve provided as arguments
to the constructor. The class defines __call__, allowing the object
to be called with the x-axis values, at which the spline should be
evaluated, returning the interpolated y-values. This is shown in
the example below for the subclass InterpolatedUnivariateSpline.
The integral,
derivatives, and
roots methods are also available
on UnivariateSpline objects, allowing definite integrals,
derivatives, and roots to be computed for the spline.
The UnivariateSpline class can also be used to smooth data by
providing a non-zero value of the smoothing parameter s, with the
same meaning as the s keyword of the splrep function
described above. This results in a spline that has fewer knots
than the number of data points, and hence is no longer strictly
an interpolating spline, but rather a smoothing spline. If this
is not desired, the InterpolatedUnivariateSpline class is available.
It is a subclass of UnivariateSpline that always passes through all
points (equivalent to forcing the smoothing parameter to 0). This
class is demonstrated in the example below.
The LSQUnivariateSpline class is the other subclass of UnivariateSpline.
It allows the user to specify the number and location of internal
knots explicitly with the parameter t. This allows for the creation
of customized splines with non-linear spacing, to interpolate in
some domains and smooth in others, or change the character of the
spline.
>>> import numpy as np
>>> import matplotlib.pyplot as plt
>>> from scipy import interpolate
InterpolatedUnivariateSpline
>>> x = np.arange(0, 2*np.pi+np.pi/4, 2*np.pi/8)
>>> y = np.sin(x)
>>> s = interpolate.InterpolatedUnivariateSpline(x, y)
>>> xnew = np.arange(0, 2*np.pi, np.pi/50)
>>> ynew = s(xnew)
>>> plt.figure()
>>> plt.plot(x, y, 'x', xnew, ynew, xnew, np.sin(xnew), x, y, 'b')
>>> plt.legend(['Linear', 'InterpolatedUnivariateSpline', 'True'])
>>> plt.axis([-0.05, 6.33, -1.05, 1.05])
>>> plt.title('InterpolatedUnivariateSpline')
>>> plt.show()
LSQUnivarateSpline with non-uniform knots
>>> t = [np.pi/2-.1, np.pi/2+.1, 3*np.pi/2-.1, 3*np.pi/2+.1]
>>> s = interpolate.LSQUnivariateSpline(x, y, t, k=2)
>>> ynew = s(xnew)
>>> plt.figure()
>>> plt.plot(x, y, 'x', xnew, ynew, xnew, np.sin(xnew), x, y, 'b')
>>> plt.legend(['Linear', 'LSQUnivariateSpline', 'True'])
>>> plt.axis([-0.05, 6.33, -1.05, 1.05])
>>> plt.title('Spline with Specified Interior Knots')
>>> plt.show()
2-D smoothing splines#
In addition to smoothing 1-D splines, the FITPACK library provides the means of fitting 2-D surfaces to two-dimensional data. The surfaces can be thought of as functions of two arguments, \(z = g(x, y)\), constructed as tensor products of 1-D splines.
Assuming that the data is held in three arrays, x, y and z,
there are two ways these data arrays can be interpreted. First—the scattered
interpolation problem—the data is assumed to be paired, i.e. the pairs of
values x[i] and y[i] represent the coordinates of the point i, which
corresponds to z[i].
The surface \(g(x, y)\) is constructed to satisfy
where \(w_i\) are non-negative weights, and s is the input parameter,
known as the smoothing factor, which controls the interplay between smoothness
of the resulting function g(x, y) and the quality of the approximation of
the data (i.e., the differences between \(g(x_i, y_i)\) and \(z_i\)). The
limit of \(s = 0\) formally corresponds to interpolation, where the surface
passes through the input data, \(g(x_i, y_i) = z_i\). See the note below however.
The second case—the rectangular grid interpolation problem—is where the data
points are assumed to be on a rectangular grid defined by all pairs of the
elements of the x and y arrays. For this problem, the z array is
assumed to be two-dimensional, and z[i, j] corresponds to (x[i], y[j]).
The bivariate spline function \(g(x, y)\) is constructed to satisfy
where s is the smoothing factor. Here the limit of \(s=0\) also
formally corresponds to interpolation, \(g(x_i, y_j) = z_{i, j}\).
Note
Internally, the smoothing surface \(g(x, y)\) is constructed by placing spline knots into the bounding box defined by the data arrays. The knots are placed automatically via the FITPACK algorithm until the desired smoothness is reached.
The knots may be placed away from the data points.
While \(s=0\) formally corresponds to a bivariate spline interpolation, the FITPACK algorithm is not meant for interpolation, and may lead to unexpected results.
For scattered data interpolation, prefer griddata; for data on a regular
grid, prefer RegularGridInterpolator.
Note
If the input data, x and y, is such that input dimensions
have incommensurate units and differ by many orders of magnitude, the
interpolant \(g(x, y)\) may have numerical artifacts. Consider
rescaling the data before interpolation.
We now consider the two spline fitting problems in turn.
Bivariate spline fitting of scattered data#
There are two interfaces for the underlying FITPACK library, a procedural one and an object-oriented interface.
Procedural interface (bisplrep)#
For (smooth) spline fitting to a 2-D surface, the function
bisplrep is available. This function takes as required inputs
the 1-D arrays x, y, and z, which represent points on the
surface \(z=f(x, y).\) The spline orders in x and y directions can
be specified via the optional parameters kx and ky. The default is
a bicubic spline, kx=ky=3.
The output of bisplrep is a list [tx ,ty, c, kx, ky] whose entries represent
respectively, the components of the knot positions, the coefficients
of the spline, and the order of the spline in each coordinate. It is
convenient to hold this list in a single object, tck, so that it can
be passed easily to the function bisplev. The
keyword, s , can be used to change the amount of smoothing performed
on the data while determining the appropriate spline. The recommended values
for \(s\) depend on the weights \(w_i\). If these are taken as \(1/d_i\),
with \(d_i\) an estimate of the standard deviation of \(z_i\), a
good value of \(s\) should be found in the range \(m- \sqrt{2m}, m +
\sqrt{2m}\), where where \(m\) is the number of data points in the x,
y, and z vectors.
The default value is \(s=m-\sqrt{2m}\). As a result, if no smoothing is desired, then ``s=0`` should be passed to `bisplrep`. (See however the note above).
To evaluate the 2-D spline and its partial derivatives
(up to the order of the spline), the function
bisplev is required. This function takes as the
first two arguments two 1-D arrays whose cross-product specifies
the domain over which to evaluate the spline. The third argument is
the tck list returned from bisplrep. If desired,
the fourth and fifth arguments provide the orders of the partial
derivative in the \(x\) and \(y\) direction, respectively.
Note
It is important to note that 2-D interpolation should not
be used to find the spline representation of images. The algorithm
used is not amenable to large numbers of input points. scipy.signal
and scipy.ndimage contain more appropriate algorithms for finding
the spline representation of an image.
The 2-D interpolation commands are intended for use when interpolating a 2-D
function as shown in the example that follows. This
example uses the mgrid command in NumPy which is
useful for defining a “mesh-grid” in many dimensions. (See also the
ogrid command if the full-mesh is not
needed). The number of output arguments and the number of dimensions
of each argument is determined by the number of indexing objects
passed in mgrid.
>>> import numpy as np
>>> from scipy import interpolate
>>> import matplotlib.pyplot as plt
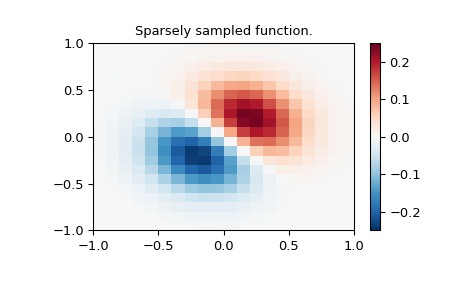
Define function over a sparse 20x20 grid
>>> x_edges, y_edges = np.mgrid[-1:1:21j, -1:1:21j]
>>> x = x_edges[:-1, :-1] + np.diff(x_edges[:2, 0])[0] / 2.
>>> y = y_edges[:-1, :-1] + np.diff(y_edges[0, :2])[0] / 2.
>>> z = (x+y) * np.exp(-6.0*(x*x+y*y))
>>> plt.figure()
>>> lims = dict(cmap='RdBu_r', vmin=-0.25, vmax=0.25)
>>> plt.pcolormesh(x_edges, y_edges, z, shading='flat', **lims)
>>> plt.colorbar()
>>> plt.title("Sparsely sampled function.")
>>> plt.show()
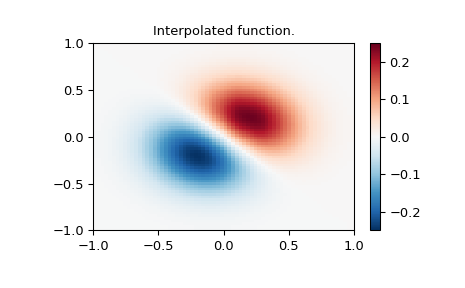
Interpolate function over a new 70x70 grid
>>> xnew_edges, ynew_edges = np.mgrid[-1:1:71j, -1:1:71j]
>>> xnew = xnew_edges[:-1, :-1] + np.diff(xnew_edges[:2, 0])[0] / 2.
>>> ynew = ynew_edges[:-1, :-1] + np.diff(ynew_edges[0, :2])[0] / 2.
>>> tck = interpolate.bisplrep(x, y, z, s=0)
>>> znew = interpolate.bisplev(xnew[:,0], ynew[0,:], tck)
>>> plt.figure()
>>> plt.pcolormesh(xnew_edges, ynew_edges, znew, shading='flat', **lims)
>>> plt.colorbar()
>>> plt.title("Interpolated function.")
>>> plt.show()
Object-oriented interface (SmoothBivariateSpline)#
The object-oriented interface for bivariate spline smoothing of scattered data,
SmoothBivariateSpline class, implements a subset of the functionality of the
bisplrep / bisplev pair, and has different defaults.
It takes the elements of the weights array equal unity, \(w_i = 1\) and constructs the knot vectors automatically given the input value of the smoothing factor s— the default value is \(m\), the number of data points.
The spline orders in the x and y directions are controlled by the optional
parameters kx and ky, with the default of kx=ky=3.
We illustrate the effect of the smoothing factor using the following example:
import numpy as np
import matplotlib.pyplot as plt
from scipy.interpolate import SmoothBivariateSpline
import warnings
warnings.simplefilter('ignore')
train_x, train_y = np.meshgrid(np.arange(-5, 5, 0.5), np.arange(-5, 5, 0.5))
train_x = train_x.flatten()
train_y = train_y.flatten()
def z_func(x, y):
return np.cos(x) + np.sin(y) ** 2 + 0.05 * x + 0.1 * y
train_z = z_func(train_x, train_y)
interp_func = SmoothBivariateSpline(train_x, train_y, train_z, s=0.0)
smth_func = SmoothBivariateSpline(train_x, train_y, train_z)
test_x = np.arange(-9, 9, 0.01)
test_y = np.arange(-9, 9, 0.01)
grid_x, grid_y = np.meshgrid(test_x, test_y)
interp_result = interp_func(test_x, test_y).T
smth_result = smth_func(test_x, test_y).T
perfect_result = z_func(grid_x, grid_y)
fig, axes = plt.subplots(1, 3, figsize=(16, 8))
extent = [test_x[0], test_x[-1], test_y[0], test_y[-1]]
opts = dict(aspect='equal', cmap='nipy_spectral', extent=extent, vmin=-1.5, vmax=2.5)
im = axes[0].imshow(perfect_result, **opts)
fig.colorbar(im, ax=axes[0], orientation='horizontal')
axes[0].plot(train_x, train_y, 'w.')
axes[0].set_title('Perfect result, sampled function', fontsize=21)
im = axes[1].imshow(smth_result, **opts)
axes[1].plot(train_x, train_y, 'w.')
fig.colorbar(im, ax=axes[1], orientation='horizontal')
axes[1].set_title('s=default', fontsize=21)
im = axes[2].imshow(interp_result, **opts)
fig.colorbar(im, ax=axes[2], orientation='horizontal')
axes[2].plot(train_x, train_y, 'w.')
axes[2].set_title('s=0', fontsize=21)
plt.tight_layout()
plt.show()
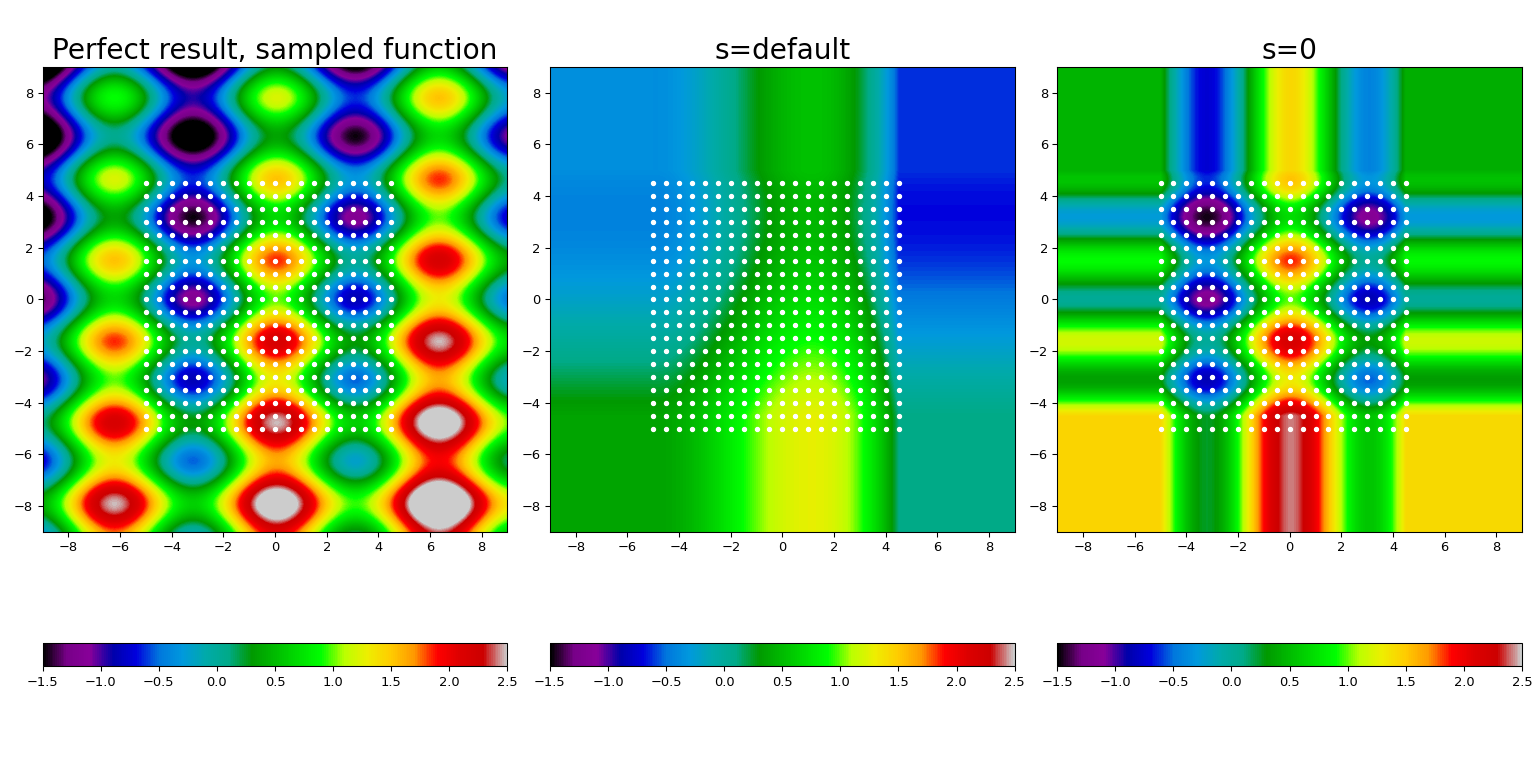
Here we take a known function (displayed at the leftmost panel), sample it on a mesh of points (shown by white dots), and construct the spline fit using the default smoothing (center panel) and forcing the interpolation (rightmost panel).
Several features are clearly visible. First, the default value of s provides
too much smoothing for this data; forcing the interpolation condition, s = 0,
allows to restore the underlying function to a reasonable accuracy. Second,
outside of the interpolation range (i.e., the area covered by
white dots) the result is extrapolated using a nearest-neighbor constant.
Finally, we had to silence the warnings (which is a bad form, yes!).
The warning here is emitted in the s=0 case, and signals an internal difficulty
FITPACK encountered when we forced the interpolation condition. If you see
this warning in your code, consider switching to bisplrep and increase its
nxest, nyest parameters (see the bisplrep docstring for more details).
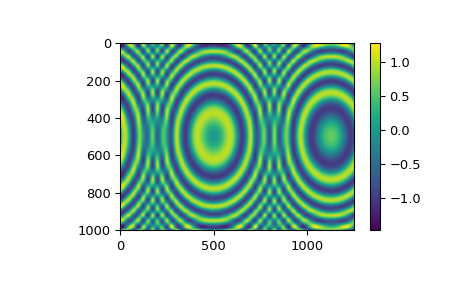
Bivariate spline fitting of data on a grid#
For gridded 2D data, fitting a smoothing tensor product spline can be done
using the RectBivariateSpline class. It has the interface similar to that of
SmoothBivariateSpline, the main difference is that the 1D input arrays x
and y are understood as definifing a 2D grid (as their outer product),
and the z array is 2D with the shape of len(x) by len(y).
The spline orders in the x and y directions are controlled by the optional
parameters kx and ky, with the default of kx=ky=3, i.e. a bicubic
spline.
The default value of the smoothing factor is s=0. We nevertheless recommend
to always specify s explicitly.
import numpy as np
import matplotlib.pyplot as plt
from scipy.interpolate import RectBivariateSpline
x = np.arange(-5.01, 5.01, 0.25) # the grid is an outer product
y = np.arange(-5.01, 7.51, 0.25) # of x and y arrays
xx, yy = np.meshgrid(x, y, indexing='ij')
z = np.sin(xx**2 + 2.*yy**2) # z array needs to be 2-D
func = RectBivariateSpline(x, y, z, s=0)
xnew = np.arange(-5.01, 5.01, 1e-2)
ynew = np.arange(-5.01, 7.51, 1e-2)
znew = func(xnew, ynew)
plt.imshow(znew)
plt.colorbar()
plt.show()
Bivariate spline fitting of data in spherical coordinates#
If your data is given in spherical coordinates, \(r = r(\theta, \phi)\),
SmoothSphereBivariateSpline and RectSphereBivariateSpline provide convenient
analogs of SmoothBivariateSpline and RectBivariateSpline, respectively.
These classes ensure the periodicity of the spline fits for \(\theta \in [0, \pi]\) and \(\phi \in [0, 2\pi]\), and offer some control over the continuity at the poles. Refer to the docstrings of these classes for details.