scipy.special.nbdtrc#
- scipy.special.nbdtrc(k, n, p, out=None) = <ufunc 'nbdtrc'>#
Negative binomial survival function.
Returns the sum of the terms k + 1 to infinity of the negative binomial distribution probability mass function,
\[F = \sum_{j=k + 1}^\infty {{n + j - 1}\choose{j}} p^n (1 - p)^j.\]In a sequence of Bernoulli trials with individual success probabilities p, this is the probability that more than k failures precede the nth success.
- Parameters:
- karray_like
The maximum number of allowed failures (nonnegative int).
- narray_like
The target number of successes (positive int).
- parray_like
Probability of success in a single event (float).
- outndarray, optional
Optional output array for the function results
- Returns:
- Fscalar or ndarray
The probability of k + 1 or more failures before n successes in a sequence of events with individual success probability p.
See also
nbdtrNegative binomial cumulative distribution function
nbdtrikNegative binomial percentile function
scipy.stats.nbinomNegative binomial distribution
Notes
If floating point values are passed for k or n, they will be truncated to integers.
The terms are not summed directly; instead the regularized incomplete beta function is employed, according to the formula,
\[\mathrm{nbdtrc}(k, n, p) = I_{1 - p}(k + 1, n).\]Wrapper for the Cephes [1] routine
nbdtrc.The negative binomial distribution is also available as
scipy.stats.nbinom. Usingnbdtrcdirectly can improve performance compared to thesfmethod ofscipy.stats.nbinom(see last example).References
[1]Cephes Mathematical Functions Library, http://www.netlib.org/cephes/
Examples
Compute the function for
k=10andn=5atp=0.5.>>> import numpy as np >>> from scipy.special import nbdtrc >>> nbdtrc(10, 5, 0.5) 0.059234619140624986
Compute the function for
n=10andp=0.5at several points by providing a NumPy array or list for k.>>> nbdtrc([5, 10, 15], 10, 0.5) array([0.84912109, 0.41190147, 0.11476147])
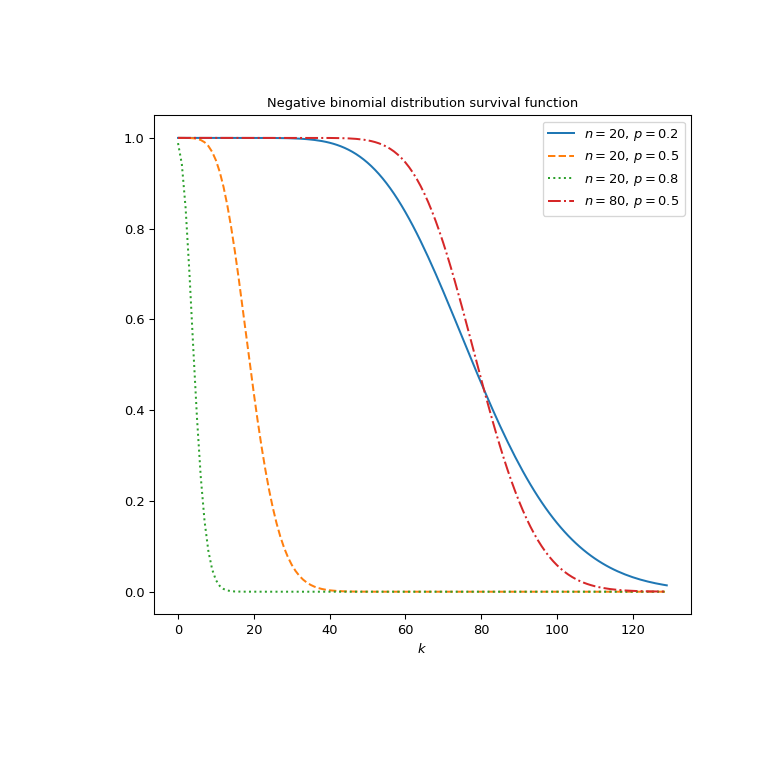
Plot the function for four different parameter sets.
>>> import matplotlib.pyplot as plt >>> k = np.arange(130) >>> n_parameters = [20, 20, 20, 80] >>> p_parameters = [0.2, 0.5, 0.8, 0.5] >>> linestyles = ['solid', 'dashed', 'dotted', 'dashdot'] >>> parameters_list = list(zip(p_parameters, n_parameters, ... linestyles)) >>> fig, ax = plt.subplots(figsize=(8, 8)) >>> for parameter_set in parameters_list: ... p, n, style = parameter_set ... nbdtrc_vals = nbdtrc(k, n, p) ... ax.plot(k, nbdtrc_vals, label=rf"$n={n},\, p={p}$", ... ls=style) >>> ax.legend() >>> ax.set_xlabel("$k$") >>> ax.set_title("Negative binomial distribution survival function") >>> plt.show()
The negative binomial distribution is also available as
scipy.stats.nbinom. Usingnbdtrcdirectly can be much faster than calling thesfmethod ofscipy.stats.nbinom, especially for small arrays or individual values. To get the same results one must use the following parametrization:nbinom(n, p).sf(k)=nbdtrc(k, n, p).>>> from scipy.stats import nbinom >>> k, n, p = 3, 5, 0.5 >>> nbdtr_res = nbdtrc(k, n, p) # this will often be faster than below >>> stats_res = nbinom(n, p).sf(k) >>> stats_res, nbdtr_res # test that results are equal (0.6367187499999999, 0.6367187499999999)
nbdtrccan evaluate different parameter sets by providing arrays with shapes compatible for broadcasting for k, n and p. Here we compute the function for three different k at four locations p, resulting in a 3x4 array.>>> k = np.array([[5], [10], [15]]) >>> p = np.array([0.3, 0.5, 0.7, 0.9]) >>> k.shape, p.shape ((3, 1), (4,))
>>> nbdtrc(k, 5, p) array([[8.49731667e-01, 3.76953125e-01, 4.73489874e-02, 1.46902600e-04], [5.15491059e-01, 5.92346191e-02, 6.72234070e-04, 9.29610100e-09], [2.37507779e-01, 5.90896606e-03, 5.55025308e-06, 3.26346760e-13]])