scipy.stats.logrank#
- scipy.stats.logrank(x, y, alternative='two-sided')[source]#
Compare the survival distributions of two samples via the logrank test.
- Parameters:
- x, yarray_like or CensoredData
Samples to compare based on their empirical survival functions.
- alternative{‘two-sided’, ‘less’, ‘greater’}, optional
Defines the alternative hypothesis.
The null hypothesis is that the survival distributions of the two groups, say X and Y, are identical.
The following alternative hypotheses [4] are available (default is ‘two-sided’):
‘two-sided’: the survival distributions of the two groups are not identical.
‘less’: survival of group X is favored: the group X failure rate function is less than the group Y failure rate function at some times.
‘greater’: survival of group Y is favored: the group X failure rate function is greater than the group Y failure rate function at some times.
- Returns:
- resLogRankResult
An object containing attributes:
- statisticfloat ndarray
The computed statistic (defined below). Its magnitude is the square root of the magnitude returned by most other logrank test implementations.
- pvaluefloat ndarray
The computed p-value of the test.
See also
Notes
The logrank test [1] compares the observed number of events to the expected number of events under the null hypothesis that the two samples were drawn from the same distribution. The statistic is
\[Z_i = \frac{\sum_{j=1}^J(O_{i,j}-E_{i,j})}{\sqrt{\sum_{j=1}^J V_{i,j}}} \rightarrow \mathcal{N}(0,1)\]where
\[E_{i,j} = O_j \frac{N_{i,j}}{N_j}, \qquad V_{i,j} = E_{i,j} \left(\frac{N_j-O_j}{N_j}\right) \left(\frac{N_j-N_{i,j}}{N_j-1}\right),\]\(i\) denotes the group (i.e. it may assume values \(x\) or \(y\), or it may be omitted to refer to the combined sample) \(j\) denotes the time (at which an event occured), \(N\) is the number of subjects at risk just before an event occured, and \(O\) is the observed number of events at that time.
The
statistic\(Z_x\) returned bylogrankis the (signed) square root of the statistic returned by many other implementations. Under the null hypothesis, \(Z_x**2\) is asymptotically distributed according to the chi-squared distribution with one degree of freedom. Consequently, \(Z_x\) is asymptotically distributed according to the standard normal distribution. The advantage of using \(Z_x\) is that the sign information (i.e. whether the observed number of events tends to be less than or greater than the number expected under the null hypothesis) is preserved, allowingscipy.stats.logrankto offer one-sided alternative hypotheses.References
[1]Mantel N. “Evaluation of survival data and two new rank order statistics arising in its consideration.” Cancer Chemotherapy Reports, 50(3):163-170, PMID: 5910392, 1966
[2]Bland, Altman, “The logrank test”, BMJ, 328:1073, DOI:10.1136/bmj.328.7447.1073, 2004
[3]“Logrank test”, Wikipedia, https://en.wikipedia.org/wiki/Logrank_test
[4]Brown, Mark. “On the choice of variance for the log rank test.” Biometrika 71.1 (1984): 65-74.
[5]Klein, John P., and Melvin L. Moeschberger. Survival analysis: techniques for censored and truncated data. Vol. 1230. New York: Springer, 2003.
Examples
Reference [2] compared the survival times of patients with two different types of recurrent malignant gliomas. The samples below record the time (number of weeks) for which each patient participated in the study. The
scipy.stats.CensoredDataclass is used because the data is right-censored: the uncensored observations correspond with observed deaths whereas the censored observations correspond with the patient leaving the study for another reason.>>> from scipy import stats >>> x = stats.CensoredData( ... uncensored=[6, 13, 21, 30, 37, 38, 49, 50, ... 63, 79, 86, 98, 202, 219], ... right=[31, 47, 80, 82, 82, 149] ... ) >>> y = stats.CensoredData( ... uncensored=[10, 10, 12, 13, 14, 15, 16, 17, 18, 20, 24, 24, ... 25, 28,30, 33, 35, 37, 40, 40, 46, 48, 76, 81, ... 82, 91, 112, 181], ... right=[34, 40, 70] ... )
We can calculate and visualize the empirical survival functions of both groups as follows.
>>> import numpy as np >>> import matplotlib.pyplot as plt >>> ax = plt.subplot() >>> ecdf_x = stats.ecdf(x) >>> ecdf_x.sf.plot(ax, label='Astrocytoma') >>> ecdf_y = stats.ecdf(y) >>> ecdf_x.sf.plot(ax, label='Glioblastoma') >>> ax.set_xlabel('Time to death (weeks)') >>> ax.set_ylabel('Empirical SF') >>> plt.legend() >>> plt.show()
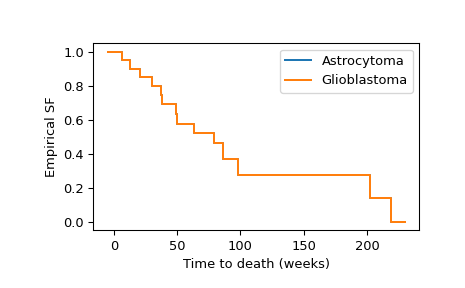
Visual inspection of the empirical survival functions suggests that the survival times tend to be different between the two groups. To formally assess whether the difference is significant at the 1% level, we use the logrank test.
>>> res = stats.logrank(x=x, y=y) >>> res.statistic -2.73799... >>> res.pvalue 0.00618...
The p-value is less than 1%, so we can consider the data to be evidence against the null hypothesis in favor of the alternative that there is a difference between the two survival functions.