scipy.signal.convolve2d¶
- scipy.signal.convolve2d(in1, in2, mode='full', boundary='fill', fillvalue=0)[source]¶
Convolve two 2-dimensional arrays.
Convolve in1 and in2 with output size determined by mode, and boundary conditions determined by boundary and fillvalue.
Parameters: in1, in2 : array_like
Two-dimensional input arrays to be convolved.
mode : str {‘full’, ‘valid’, ‘same’}, optional
A string indicating the size of the output:
- full
The output is the full discrete linear convolution of the inputs. (Default)
- valid
The output consists only of those elements that do not rely on the zero-padding.
- same
The output is the same size as in1, centered with respect to the ‘full’ output.
boundary : str {‘fill’, ‘wrap’, ‘symm’}, optional
A flag indicating how to handle boundaries:
- fill
pad input arrays with fillvalue. (default)
- wrap
circular boundary conditions.
- symm
symmetrical boundary conditions.
fillvalue : scalar, optional
Value to fill pad input arrays with. Default is 0.
Returns: out : ndarray
A 2-dimensional array containing a subset of the discrete linear convolution of in1 with in2.
Examples
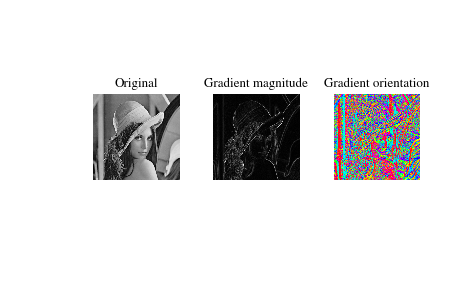
Compute the gradient of an image by 2D convolution with a complex Scharr operator. (Horizontal operator is real, vertical is imaginary.) Use symmetric boundary condition to avoid creating edges at the image boundaries.
>>> from scipy import signal >>> from scipy import misc >>> lena = misc.lena() >>> scharr = np.array([[ -3-3j, 0-10j, +3 -3j], ... [-10+0j, 0+ 0j, +10 +0j], ... [ -3+3j, 0+10j, +3 +3j]]) # Gx + j*Gy >>> grad = signal.convolve2d(lena, scharr, boundary='symm', mode='same')
>>> import matplotlib.pyplot as plt >>> fig, (ax_orig, ax_mag, ax_ang) = plt.subplots(1, 3) >>> ax_orig.imshow(lena, cmap='gray') >>> ax_orig.set_title('Original') >>> ax_orig.set_axis_off() >>> ax_mag.imshow(np.absolute(grad), cmap='gray') >>> ax_mag.set_title('Gradient magnitude') >>> ax_mag.set_axis_off() >>> ax_ang.imshow(np.angle(grad), cmap='hsv') # hsv is cyclic, like angles >>> ax_ang.set_title('Gradient orientation') >>> ax_ang.set_axis_off() >>> fig.show()