scipy.stats.fit#
- scipy.stats.fit(dist, data, bounds=None, *, guess=None, optimizer=<function differential_evolution>)[source]#
Fit a discrete or continuous distribution to data
Given a distribution, data, and bounds on the parameters of the distribution, return maximum likelihood estimates of the parameters.
- Parameters
- dist
scipy.stats.rv_continuousorscipy.stats.rv_discrete The object representing the distribution to be fit to the data.
- data1D array_like
The data to which the distribution is to be fit. If the data contain any of
np.nan,np.inf, or -np.inf, the fit method will raise aValueError.- boundsdict or sequence of tuples, optional
If a dictionary, each key is the name of a parameter of the distribution, and the corresponding value is a tuple containing the lower and upper bound on that parameter. If the distribution is defined only for a finite range of values of that parameter, no entry for that parameter is required; e.g., some distributions have parameters which must be on the interval [0, 1]. Bounds for parameters location (
loc) and scale (scale) are optional; by default, they are fixed to 0 and 1, respectively.If a sequence, element i is a tuple containing the lower and upper bound on the ith parameter of the distribution. In this case, bounds for all distribution shape parameters must be provided. Optionally, bounds for location and scale may follow the distribution shape parameters.
If a shape is to be held fixed (e.g. if it is known), the lower and upper bounds may be equal. If a user-provided lower or upper bound is beyond a bound of the domain for which the distribution is defined, the bound of the distribution’s domain will replace the user-provided value. Similarly, parameters which must be integral will be constrained to integral values within the user-provided bounds.
- guessdict or array_like, optional
If a dictionary, each key is the name of a parameter of the distribution, and the corresponding value is a guess for the value of the parameter.
If a sequence, element i is a guess for the ith parameter of the distribution. In this case, guesses for all distribution shape parameters must be provided.
If guess is not provided, guesses for the decision variables will not be passed to the optimizer. If guess is provided, guesses for any missing parameters will be set at the mean of the lower and upper bounds. Guesses for parameters which must be integral will be rounded to integral values, and guesses that lie outside the intersection of the user-provided bounds and the domain of the distribution will be clipped.
- optimizercallable, optional
optimizer is a callable that accepts the following positional argument.
- funcallable
The objective function to be optimized. fun accepts one argument
x, candidate shape parameters of the distribution, and returns the negative log-likelihood function givenx, dist, and the provided data. The job of optimizer is to find values of the decision variables that minimizes fun.
optimizer must also accepts the following keyword argument.
- boundssequence of tuples
The bounds on values of the decision variables; each element will be a tuple containing the lower and upper bound on a decision variable.
If guess is provided, optimizer must also accept the following keyword argument.
- x0array_like
The guesses for each decision variable.
If the distribution has any shape parameters that must be integral or if the distribution is discrete and the location parameter is not fixed, optimizer must also accept the following keyword argument.
- integralityarray_like of bools
For each decision variable, True if the decision variable is must be constrained to integer values and False if the decision variable is continuous.
optimizer must return an object, such as an instance of
scipy.optimize.OptimizeResult, which holds the optimal values of the decision variables in an attributex. If attributesfun,status, ormessageare provided, they will be included in the result object returned byfit.
- dist
- Returns
- result
FitResult An object with the following fields.
- paramsnamedtuple
A namedtuple containing the maximum likelihood estimates of the shape parameters, location, and (if applicable) scale of the distribution.
- successbool or None
Whether the optimizer considered the optimization to terminate successfully or not.
- messagestr or None
Any status message provided by the optimizer.
The object has the following method:
- nllf(params=None, data=None)
By default, the negative log-likehood function at the fitted params for the given data. Accepts a tuple containing alternative shapes, location, and scale of the distribution and an array of alternative data.
- plot(ax=None)
Superposes the PDF/PMF of the fitted distribution over a normalized histogram of the data.
- result
See also
Notes
Optimization is more likely to converge to the maximum likelihood estimate when the user provides tight bounds containing the maximum likelihood estimate. For example, when fitting a binomial distribution to data, the number of experiments underlying each sample may be known, in which case the corresponding shape parameter
ncan be fixed.Examples
Suppose we wish to fit a distribution to the following data.
>>> import numpy as np >>> from scipy import stats >>> rng = np.random.default_rng() >>> dist = stats.nbinom >>> shapes = (5, 0.5) >>> data = dist.rvs(*shapes, size=1000, random_state=rng)
Suppose we do not know how the data were generated, but we suspect that it follows a negative binomial distribution with parameters n and p. (See
scipy.stats.nbinom.) We believe that the parameter n was fewer than 30, and we know that the parameter p must lie on the interval [0, 1]. We record this information in a variable bounds and pass this information tofit.>>> bounds = [(0, 30), (0, 1)] >>> res = stats.fit(dist, data, bounds)
fitsearches within the user-specified bounds for the values that best match the data (in the sense of maximum likelihood estimation). In this case, it found shape values similar to those from which the data were actually generated.>>> res.params FitParams(n=5.0, p=0.5028157644634368, loc=0.0) # may vary
We can visualize the results by superposing the probability mass function of the distribution (with the shapes fit to the data) over a normalized histogram of the data.
>>> import matplotlib.pyplot as plt # matplotlib must be installed to plot >>> res.plot() >>> plt.show()
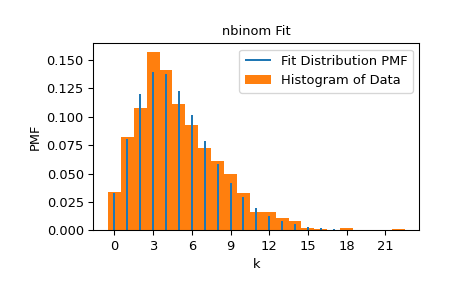
Note that the estimate for n was exactly integral; this is because the domain of the
nbinomPMF includes only integral n, and thenbinomobject “knows” that.nbinomalso knows that the shape p must be a value between 0 and 1. In such a case - when the domain of the distribution with respect to a parameter is finite - we are not required to specify bounds for the parameter.>>> bounds = {'n': (0, 30)} # omit parameter p using a `dict` >>> res2 = stats.fit(dist, data, bounds) >>> res2.params FitParams(n=5.0, p=0.5016492009232932, loc=0.0) # may vary
If we wish to force the distribution to be fit with n fixed at 6, we can set both the lower and upper bounds on n to 6. Note, however, that the value of the objective function being optimized is typically worse (higher) in this case.
>>> bounds = {'n': (6, 6)} # fix parameter `n` >>> res3 = stats.fit(dist, data, bounds) >>> res3.params FitParams(n=6.0, p=0.5486556076755706, loc=0.0) # may vary >>> res3.nllf() > res.nllf() True # may vary
Note that the numerical results of the previous examples are typical, but they may vary because the default optimizer used by
fit,scipy.optimize.differential_evolution, is stochastic. However, we can customize the settings used by the optimizer to ensure reproducibility - or even use a different optimizer entirely - using the optimizer parameter.>>> from scipy.optimize import differential_evolution >>> rng = np.random.default_rng(767585560716548) >>> def optimizer(fun, bounds, *, integrality): ... return differential_evolution(fun, bounds, strategy='best2bin', ... seed=rng, integrality=integrality) >>> bounds = [(0, 30), (0, 1)] >>> res4 = stats.fit(dist, data, bounds, optimizer=optimizer) >>> res4.params FitParams(n=5.0, p=0.5015183149259951, loc=0.0)