scipy.stats.spearmanr#
- scipy.stats.spearmanr(a, b=None, axis=0, nan_policy='propagate', alternative='two-sided')[source]#
Calculate a Spearman correlation coefficient with associated p-value.
The Spearman rank-order correlation coefficient is a nonparametric measure of the monotonicity of the relationship between two datasets. Like other correlation coefficients, this one varies between -1 and +1 with 0 implying no correlation. Correlations of -1 or +1 imply an exact monotonic relationship. Positive correlations imply that as x increases, so does y. Negative correlations imply that as x increases, y decreases.
The p-value roughly indicates the probability of an uncorrelated system producing datasets that have a Spearman correlation at least as extreme as the one computed from these datasets. Although calculation of the p-value does not make strong assumptions about the distributions underlying the samples, it is only accurate for very large samples (>500 observations). For smaller sample sizes, consider a permutation test (see Examples section below).
- Parameters:
- a, b1D or 2D array_like, b is optional
One or two 1-D or 2-D arrays containing multiple variables and observations. When these are 1-D, each represents a vector of observations of a single variable. For the behavior in the 2-D case, see under
axis, below. Both arrays need to have the same length in theaxisdimension.- axisint or None, optional
If axis=0 (default), then each column represents a variable, with observations in the rows. If axis=1, the relationship is transposed: each row represents a variable, while the columns contain observations. If axis=None, then both arrays will be raveled.
- nan_policy{‘propagate’, ‘raise’, ‘omit’}, optional
Defines how to handle when input contains nan. The following options are available (default is ‘propagate’):
‘propagate’: returns nan
‘raise’: throws an error
‘omit’: performs the calculations ignoring nan values
- alternative{‘two-sided’, ‘less’, ‘greater’}, optional
Defines the alternative hypothesis. Default is ‘two-sided’. The following options are available:
‘two-sided’: the correlation is nonzero
‘less’: the correlation is negative (less than zero)
‘greater’: the correlation is positive (greater than zero)
Added in version 1.7.0.
- Returns:
- resSignificanceResult
An object containing attributes:
- statisticfloat or ndarray (2-D square)
Spearman correlation matrix or correlation coefficient (if only 2 variables are given as parameters). Correlation matrix is square with length equal to total number of variables (columns or rows) in
aandbcombined.- pvaluefloat
The p-value for a hypothesis test whose null hypothesis is that two samples have no ordinal correlation. See alternative above for alternative hypotheses. pvalue has the same shape as statistic.
- Warns:
ConstantInputWarningRaised if an input is a constant array. The correlation coefficient is not defined in this case, so
np.nanis returned.
References
[1]Zwillinger, D. and Kokoska, S. (2000). CRC Standard Probability and Statistics Tables and Formulae. Chapman & Hall: New York. 2000. Section 14.7
[2]Kendall, M. G. and Stuart, A. (1973). The Advanced Theory of Statistics, Volume 2: Inference and Relationship. Griffin. 1973. Section 31.18
[3]Kershenobich, D., Fierro, F. J., & Rojkind, M. (1970). The relationship between the free pool of proline and collagen content in human liver cirrhosis. The Journal of Clinical Investigation, 49(12), 2246-2249.
[4]Hollander, M., Wolfe, D. A., & Chicken, E. (2013). Nonparametric statistical methods. John Wiley & Sons.
[5]B. Phipson and G. K. Smyth. “Permutation P-values Should Never Be Zero: Calculating Exact P-values When Permutations Are Randomly Drawn.” Statistical Applications in Genetics and Molecular Biology 9.1 (2010).
[6]Ludbrook, J., & Dudley, H. (1998). Why permutation tests are superior to t and F tests in biomedical research. The American Statistician, 52(2), 127-132.
Examples
Consider the following data from [3], which studied the relationship between free proline (an amino acid) and total collagen (a protein often found in connective tissue) in unhealthy human livers.
The
xandyarrays below record measurements of the two compounds. The observations are paired: each free proline measurement was taken from the same liver as the total collagen measurement at the same index.>>> import numpy as np >>> # total collagen (mg/g dry weight of liver) >>> x = np.array([7.1, 7.1, 7.2, 8.3, 9.4, 10.5, 11.4]) >>> # free proline (μ mole/g dry weight of liver) >>> y = np.array([2.8, 2.9, 2.8, 2.6, 3.5, 4.6, 5.0])
These data were analyzed in [4] using Spearman’s correlation coefficient, a statistic sensitive to monotonic correlation between the samples.
>>> from scipy import stats >>> res = stats.spearmanr(x, y) >>> res.statistic 0.7000000000000001
The value of this statistic tends to be high (close to 1) for samples with a strongly positive ordinal correlation, low (close to -1) for samples with a strongly negative ordinal correlation, and small in magnitude (close to zero) for samples with weak ordinal correlation.
The test is performed by comparing the observed value of the statistic against the null distribution: the distribution of statistic values derived under the null hypothesis that total collagen and free proline measurements are independent.
For this test, the statistic can be transformed such that the null distribution for large samples is Student’s t distribution with
len(x) - 2degrees of freedom.>>> import matplotlib.pyplot as plt >>> dof = len(x)-2 # len(x) == len(y) >>> dist = stats.t(df=dof) >>> t_vals = np.linspace(-5, 5, 100) >>> pdf = dist.pdf(t_vals) >>> fig, ax = plt.subplots(figsize=(8, 5)) >>> def plot(ax): # we'll reuse this ... ax.plot(t_vals, pdf) ... ax.set_title("Spearman's Rho Test Null Distribution") ... ax.set_xlabel("statistic") ... ax.set_ylabel("probability density") >>> plot(ax) >>> plt.show()
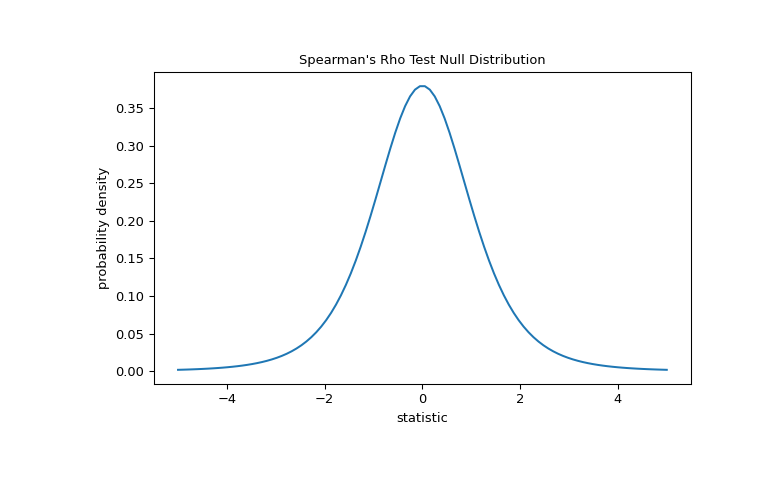
The comparison is quantified by the p-value: the proportion of values in the null distribution as extreme or more extreme than the observed value of the statistic. In a two-sided test in which the statistic is positive, elements of the null distribution greater than the transformed statistic and elements of the null distribution less than the negative of the observed statistic are both considered “more extreme”.
>>> fig, ax = plt.subplots(figsize=(8, 5)) >>> plot(ax) >>> rs = res.statistic # original statistic >>> transformed = rs * np.sqrt(dof / ((rs+1.0)*(1.0-rs))) >>> pvalue = dist.cdf(-transformed) + dist.sf(transformed) >>> annotation = (f'p-value={pvalue:.4f}\n(shaded area)') >>> props = dict(facecolor='black', width=1, headwidth=5, headlength=8) >>> _ = ax.annotate(annotation, (2.7, 0.025), (3, 0.03), arrowprops=props) >>> i = t_vals >= transformed >>> ax.fill_between(t_vals[i], y1=0, y2=pdf[i], color='C0') >>> i = t_vals <= -transformed >>> ax.fill_between(t_vals[i], y1=0, y2=pdf[i], color='C0') >>> ax.set_xlim(-5, 5) >>> ax.set_ylim(0, 0.1) >>> plt.show()
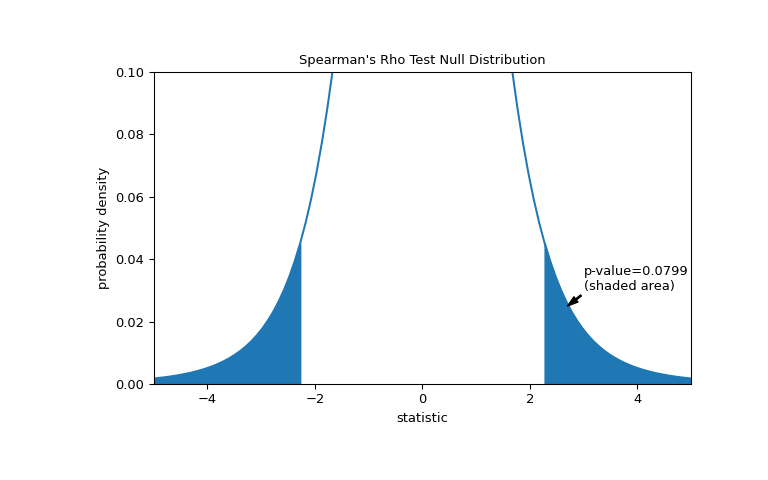
>>> res.pvalue 0.07991669030889909 # two-sided p-value
If the p-value is “small” - that is, if there is a low probability of sampling data from independent distributions that produces such an extreme value of the statistic - this may be taken as evidence against the null hypothesis in favor of the alternative: the distribution of total collagen and free proline are not independent. Note that:
The inverse is not true; that is, the test is not used to provide evidence for the null hypothesis.
The threshold for values that will be considered “small” is a choice that should be made before the data is analyzed [5] with consideration of the risks of both false positives (incorrectly rejecting the null hypothesis) and false negatives (failure to reject a false null hypothesis).
Small p-values are not evidence for a large effect; rather, they can only provide evidence for a “significant” effect, meaning that they are unlikely to have occurred under the null hypothesis.
Suppose that before performing the experiment, the authors had reason to predict a positive correlation between the total collagen and free proline measurements, and that they had chosen to assess the plausibility of the null hypothesis against a one-sided alternative: free proline has a positive ordinal correlation with total collagen. In this case, only those values in the null distribution that are as great or greater than the observed statistic are considered to be more extreme.
>>> res = stats.spearmanr(x, y, alternative='greater') >>> res.statistic 0.7000000000000001 # same statistic >>> fig, ax = plt.subplots(figsize=(8, 5)) >>> plot(ax) >>> pvalue = dist.sf(transformed) >>> annotation = (f'p-value={pvalue:.6f}\n(shaded area)') >>> props = dict(facecolor='black', width=1, headwidth=5, headlength=8) >>> _ = ax.annotate(annotation, (3, 0.018), (3.5, 0.03), arrowprops=props) >>> i = t_vals >= transformed >>> ax.fill_between(t_vals[i], y1=0, y2=pdf[i], color='C0') >>> ax.set_xlim(1, 5) >>> ax.set_ylim(0, 0.1) >>> plt.show()
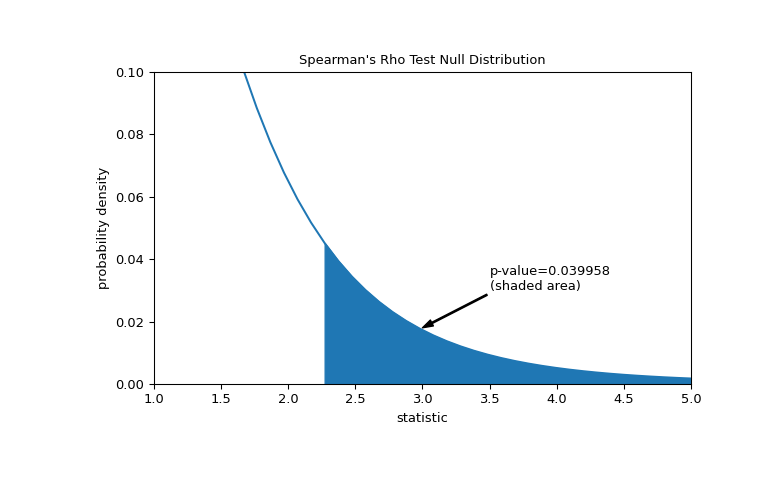
>>> res.pvalue 0.03995834515444954 # one-sided p-value; half of the two-sided p-value
Note that the t-distribution provides an asymptotic approximation of the null distribution; it is only accurate for samples with many observations. For small samples, it may be more appropriate to perform a permutation test: Under the null hypothesis that total collagen and free proline are independent, each of the free proline measurements were equally likely to have been observed with any of the total collagen measurements. Therefore, we can form an exact null distribution by calculating the statistic under each possible pairing of elements between
xandy.>>> def statistic(x): # explore all possible pairings by permuting `x` ... rs = stats.spearmanr(x, y).statistic # ignore pvalue ... transformed = rs * np.sqrt(dof / ((rs+1.0)*(1.0-rs))) ... return transformed >>> ref = stats.permutation_test((x,), statistic, alternative='greater', ... permutation_type='pairings') >>> fig, ax = plt.subplots(figsize=(8, 5)) >>> plot(ax) >>> ax.hist(ref.null_distribution, np.linspace(-5, 5, 26), ... density=True) >>> ax.legend(['aymptotic approximation\n(many observations)', ... f'exact \n({len(ref.null_distribution)} permutations)']) >>> plt.show()
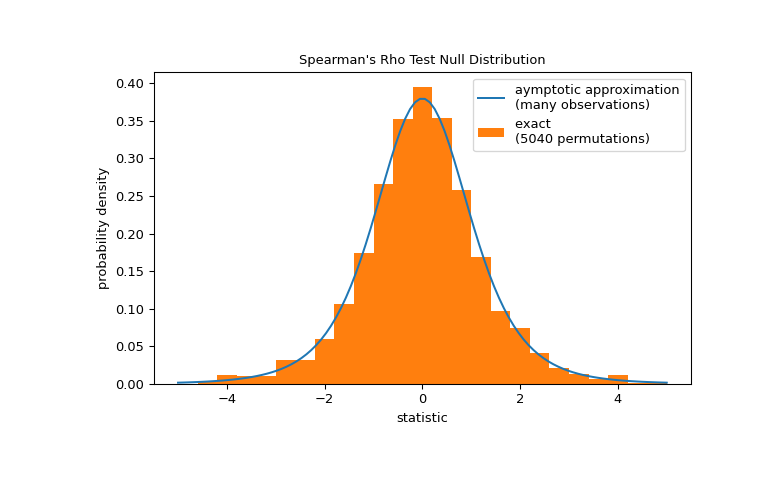
>>> ref.pvalue 0.04563492063492063 # exact one-sided p-value