scipy.stats.qmc.geometric_discrepancy#
- scipy.stats.qmc.geometric_discrepancy(sample, method='mindist', metric='euclidean')[source]#
Discrepancy of a given sample based on its geometric properties.
- Parameters:
- samplearray_like (n, d)
The sample to compute the discrepancy from.
- method{“mindist”, “mst”}, optional
The method to use. One of
mindistfor minimum distance (default) ormstfor minimum spanning tree.- metricstr or callable, optional
The distance metric to use. See the documentation for
scipy.spatial.distance.pdistfor the available metrics and the default.
- Returns:
- discrepancyfloat
Discrepancy (higher values correspond to greater sample uniformity).
See also
Notes
The discrepancy can serve as a simple measure of quality of a random sample. This measure is based on the geometric properties of the distribution of points in the sample, such as the minimum distance between any pair of points, or the mean edge length in a minimum spanning tree.
The higher the value is, the better the coverage of the parameter space is. Note that this is different from
scipy.stats.qmc.discrepancy, where lower values correspond to higher quality of the sample.Also note that when comparing different sampling strategies using this function, the sample size must be kept constant.
It is possible to calculate two metrics from the minimum spanning tree: the mean edge length and the standard deviation of edges lengths. Using both metrics offers a better picture of uniformity than either metric alone, with higher mean and lower standard deviation being preferable (see [1] for a brief discussion). This function currently only calculates the mean edge length.
References
[1]Franco J. et al. “Minimum Spanning Tree: A new approach to assess the quality of the design of computer experiments.” Chemometrics and Intelligent Laboratory Systems, 97 (2), pp. 164-169, 2009.
Examples
Calculate the quality of the sample using the minimum euclidean distance (the defaults):
>>> import numpy as np >>> from scipy.stats import qmc >>> rng = np.random.default_rng() >>> sample = qmc.LatinHypercube(d=2, seed=rng).random(50) >>> qmc.geometric_discrepancy(sample) 0.03708161435687876
Calculate the quality using the mean edge length in the minimum spanning tree:
>>> qmc.geometric_discrepancy(sample, method='mst') 0.1105149978798376
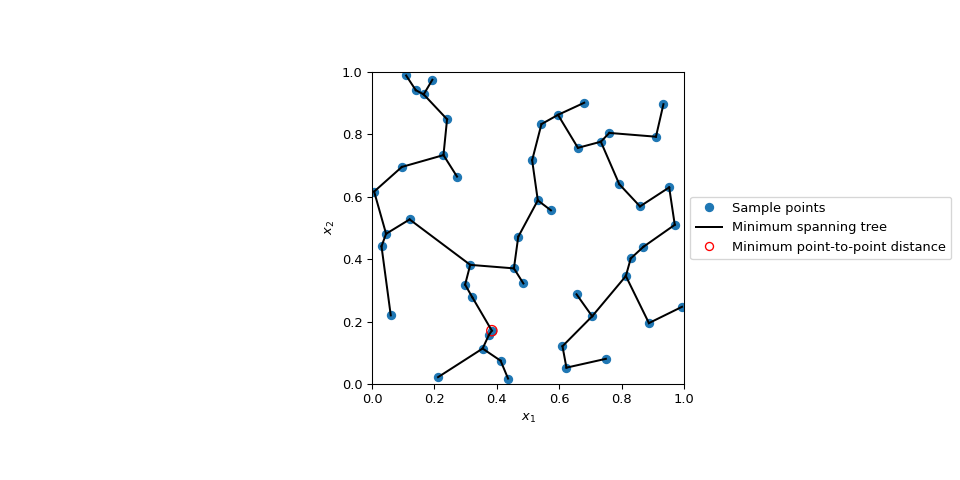
Display the minimum spanning tree and the points with the smallest distance:
>>> import matplotlib.pyplot as plt >>> from matplotlib.lines import Line2D >>> from scipy.sparse.csgraph import minimum_spanning_tree >>> from scipy.spatial.distance import pdist, squareform >>> dist = pdist(sample) >>> mst = minimum_spanning_tree(squareform(dist)) >>> edges = np.where(mst.toarray() > 0) >>> edges = np.asarray(edges).T >>> min_dist = np.min(dist) >>> min_idx = np.argwhere(squareform(dist) == min_dist)[0] >>> fig, ax = plt.subplots(figsize=(10, 5)) >>> _ = ax.set(aspect='equal', xlabel=r'$x_1$', ylabel=r'$x_2$', ... xlim=[0, 1], ylim=[0, 1]) >>> for edge in edges: ... ax.plot(sample[edge, 0], sample[edge, 1], c='k') >>> ax.scatter(sample[:, 0], sample[:, 1]) >>> ax.add_patch(plt.Circle(sample[min_idx[0]], min_dist, color='red', fill=False)) >>> markers = [ ... Line2D([0], [0], marker='o', lw=0, label='Sample points'), ... Line2D([0], [0], color='k', label='Minimum spanning tree'), ... Line2D([0], [0], marker='o', lw=0, markerfacecolor='w', markeredgecolor='r', ... label='Minimum point-to-point distance'), ... ] >>> ax.legend(handles=markers, loc='center left', bbox_to_anchor=(1, 0.5)); >>> plt.show()