scipy.interpolate.SmoothBivariateSpline.ev#
- SmoothBivariateSpline.ev(xi, yi, dx=0, dy=0)[source]#
Evaluate the spline at points
Returns the interpolated value at
(xi[i], yi[i]), i=0,...,len(xi)-1.- Parameters:
- xi, yiarray_like
Input coordinates. Standard Numpy broadcasting is obeyed. The ordering of axes is consistent with
np.meshgrid(..., indexing="ij")and inconsistent with the default orderingnp.meshgrid(..., indexing="xy").- dxint, optional
Order of x-derivative
New in version 0.14.0.
- dyint, optional
Order of y-derivative
New in version 0.14.0.
Examples
Suppose that we want to bilinearly interpolate an exponentially decaying function in 2 dimensions.
>>> import numpy as np >>> from scipy.interpolate import RectBivariateSpline >>> def f(x, y): ... return np.exp(-np.sqrt((x / 2) ** 2 + y**2))
We sample the function on a coarse grid and set up the interpolator. Note that the default
indexing="xy"of meshgrid would result in an unexpected (transposed) result after interpolation.>>> xarr = np.linspace(-3, 3, 21) >>> yarr = np.linspace(-3, 3, 21) >>> xgrid, ygrid = np.meshgrid(xarr, yarr, indexing="ij") >>> zdata = f(xgrid, ygrid) >>> rbs = RectBivariateSpline(xarr, yarr, zdata, kx=1, ky=1)
Next we sample the function along a diagonal slice through the coordinate space on a finer grid using interpolation.
>>> xinterp = np.linspace(-3, 3, 201) >>> yinterp = np.linspace(3, -3, 201) >>> zinterp = rbs.ev(xinterp, yinterp)
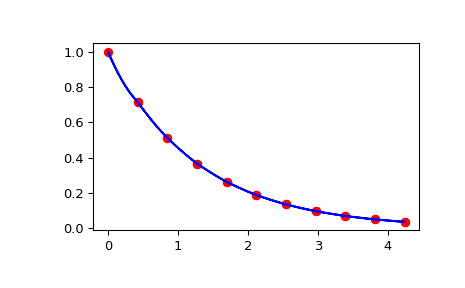
And check that the interpolation passes through the function evaluations as a function of the distance from the origin along the slice.
>>> import matplotlib.pyplot as plt >>> fig = plt.figure() >>> ax1 = fig.add_subplot(1, 1, 1) >>> ax1.plot(np.sqrt(xarr**2 + yarr**2), np.diag(zdata), "or") >>> ax1.plot(np.sqrt(xinterp**2 + yinterp**2), zinterp, "-b") >>> plt.show()