scipy.stats.kurtosistest#
- scipy.stats.kurtosistest(a, axis=0, nan_policy='propagate', alternative='two-sided')[source]#
Test whether a dataset has normal kurtosis.
This function tests the null hypothesis that the kurtosis of the population from which the sample was drawn is that of the normal distribution.
- Parameters:
- aarray
Array of the sample data.
- axisint or None, optional
Axis along which to compute test. Default is 0. If None, compute over the whole array a.
- nan_policy{‘propagate’, ‘raise’, ‘omit’}, optional
Defines how to handle when input contains nan. The following options are available (default is ‘propagate’):
‘propagate’: returns nan
‘raise’: throws an error
‘omit’: performs the calculations ignoring nan values
- alternative{‘two-sided’, ‘less’, ‘greater’}, optional
Defines the alternative hypothesis. The following options are available (default is ‘two-sided’):
‘two-sided’: the kurtosis of the distribution underlying the sample is different from that of the normal distribution
‘less’: the kurtosis of the distribution underlying the sample is less than that of the normal distribution
‘greater’: the kurtosis of the distribution underlying the sample is greater than that of the normal distribution
New in version 1.7.0.
- Returns:
- statisticfloat
The computed z-score for this test.
- pvaluefloat
The p-value for the hypothesis test.
Notes
Valid only for n>20. This function uses the method described in [1].
References
[1] (1,2)see e.g. F. J. Anscombe, W. J. Glynn, “Distribution of the kurtosis statistic b2 for normal samples”, Biometrika, vol. 70, pp. 227-234, 1983.
[2]Shapiro, S. S., & Wilk, M. B. (1965). An analysis of variance test for normality (complete samples). Biometrika, 52(3/4), 591-611.
[3]B. Phipson and G. K. Smyth. “Permutation P-values Should Never Be Zero: Calculating Exact P-values When Permutations Are Randomly Drawn.” Statistical Applications in Genetics and Molecular Biology 9.1 (2010).
[4]Panagiotakos, D. B. (2008). The value of p-value in biomedical research. The open cardiovascular medicine journal, 2, 97.
Examples
Suppose we wish to infer from measurements whether the weights of adult human males in a medical study are not normally distributed [2]. The weights (lbs) are recorded in the array
xbelow.>>> import numpy as np >>> x = np.array([148, 154, 158, 160, 161, 162, 166, 170, 182, 195, 236])
The kurtosis test from [1] begins by computing a statistic based on the sample (excess/Fisher) kurtosis.
>>> from scipy import stats >>> res = stats.kurtosistest(x) >>> res.statistic 2.3048235214240873
(The test warns that our sample has too few observations to perform the test. We’ll return to this at the end of the example.) Because normal distributions have zero excess kurtosis (by definition), the magnitude of this statistic tends to be low for samples drawn from a normal distribution.
The test is performed by comparing the observed value of the statistic against the null distribution: the distribution of statistic values derived under the null hypothesis that the weights were drawn from a normal distribution.
For this test, the null distribution of the statistic for very large samples is the standard normal distribution.
>>> import matplotlib.pyplot as plt >>> dist = stats.norm() >>> kt_val = np.linspace(-5, 5, 100) >>> pdf = dist.pdf(kt_val) >>> fig, ax = plt.subplots(figsize=(8, 5)) >>> def kt_plot(ax): # we'll reuse this ... ax.plot(kt_val, pdf) ... ax.set_title("Kurtosis Test Null Distribution") ... ax.set_xlabel("statistic") ... ax.set_ylabel("probability density") >>> kt_plot(ax) >>> plt.show()
The comparison is quantified by the p-value: the proportion of values in the null distribution as extreme or more extreme than the observed value of the statistic. In a two-sided test in which the statistic is positive, elements of the null distribution greater than the observed statistic and elements of the null distribution less than the negative of the observed statistic are both considered “more extreme”.
>>> fig, ax = plt.subplots(figsize=(8, 5)) >>> kt_plot(ax) >>> pvalue = dist.cdf(-res.statistic) + dist.sf(res.statistic) >>> annotation = (f'p-value={pvalue:.3f}\n(shaded area)') >>> props = dict(facecolor='black', width=1, headwidth=5, headlength=8) >>> _ = ax.annotate(annotation, (3, 0.005), (3.25, 0.02), arrowprops=props) >>> i = kt_val >= res.statistic >>> ax.fill_between(kt_val[i], y1=0, y2=pdf[i], color='C0') >>> i = kt_val <= -res.statistic >>> ax.fill_between(kt_val[i], y1=0, y2=pdf[i], color='C0') >>> ax.set_xlim(-5, 5) >>> ax.set_ylim(0, 0.1) >>> plt.show()
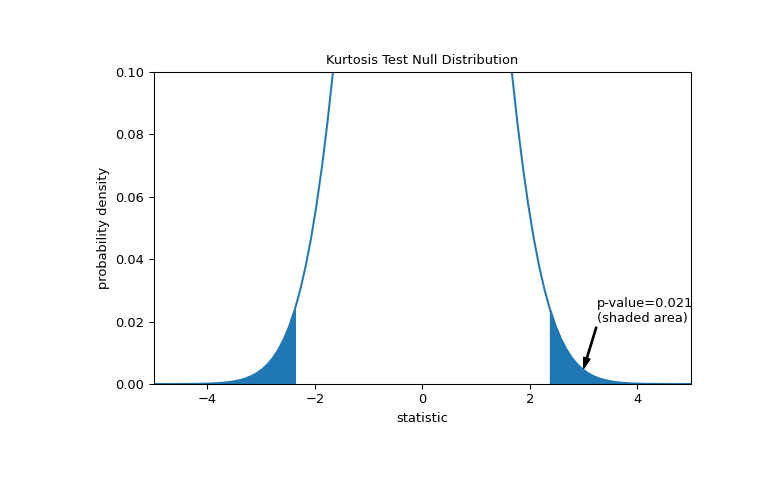
>>> res.pvalue 0.0211764592113868
If the p-value is “small” - that is, if there is a low probability of sampling data from a normally distributed population that produces such an extreme value of the statistic - this may be taken as evidence against the null hypothesis in favor of the alternative: the weights were not drawn from a normal distribution. Note that:
The inverse is not true; that is, the test is not used to provide evidence for the null hypothesis.
The threshold for values that will be considered “small” is a choice that should be made before the data is analyzed [3] with consideration of the risks of both false positives (incorrectly rejecting the null hypothesis) and false negatives (failure to reject a false null hypothesis).
Note that the standard normal distribution provides an asymptotic approximation of the null distribution; it is only accurate for samples with many observations. This is the reason we received a warning at the beginning of the example; our sample is quite small. In this case,
scipy.stats.monte_carlo_testmay provide a more accurate, albeit stochastic, approximation of the exact p-value.>>> def statistic(x, axis): ... # get just the skewtest statistic; ignore the p-value ... return stats.kurtosistest(x, axis=axis).statistic >>> res = stats.monte_carlo_test(x, stats.norm.rvs, statistic) >>> fig, ax = plt.subplots(figsize=(8, 5)) >>> kt_plot(ax) >>> ax.hist(res.null_distribution, np.linspace(-5, 5, 50), ... density=True) >>> ax.legend(['aymptotic approximation\n(many observations)', ... 'Monte Carlo approximation\n(11 observations)']) >>> plt.show()
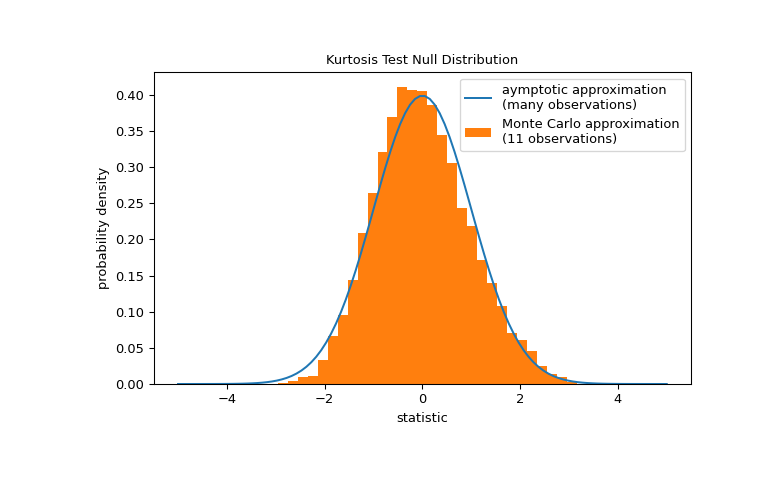
>>> res.pvalue 0.0272 # may vary
Furthermore, despite their stochastic nature, p-values computed in this way can be used to exactly control the rate of false rejections of the null hypothesis [4].