Discrete Alias Urn (DAU)#
Required: probability vector (PV) or the PMF along with a finite domain
Speed:
Set-up: slow (linear with the vector-length)
Sampling: very fast
DAU samples from distributions with arbitrary but finite probability vectors (PV) of length N. The algorithm is based on an ingenious method by A.J. Walker and requires a table of size (at least) N. It needs one random number and only one comparison for each generated random variate. The setup time for constructing the tables is O(N).
>>> import numpy as np
>>> from scipy.stats.sampling import DiscreteAliasUrn
>>>
>>> pv = [0.18, 0.02, 0.8]
>>> urng = np.random.default_rng()
>>> rng = DiscreteAliasUrn(pv, random_state=urng)
>>> rng.rvs()
0
By default, the probability vector is indexed starting at 0. However, this
can be changed by passing a domain parameter. When domain is given
in combination with the PV, it has the effect of relocating the
distribution from (0, len(pv)) to (domain[0], domain[0] + len(pv)).
domain[1] is ignored in this case.
>>> rng = DiscreteAliasUrn(pv, domain=(10, 13), random_state=urng)
>>> rng.rvs()
12
The method also works when no probability vector but a PMF is given.
In that case, a bounded (finite) domain must also be given either by
passing the domain parameter explicitly or by providing a support
method in the distribution object:
>>> class Distribution:
... def __init__(self, c):
... self.c = c
... def pmf(self, x):
... return x**self.c
... def support(self):
... return (0, 10)
...
>>> dist = Distribution(2)
>>> rng = DiscreteAliasUrn(dist, random_state=urng)
>>> rng.rvs()
10
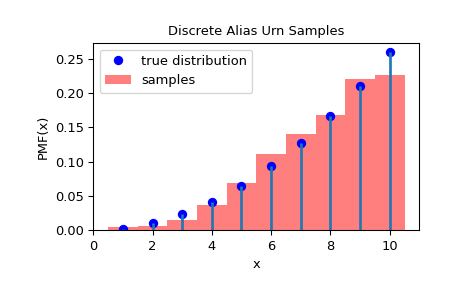
>>> import matplotlib.pyplot as plt
>>> from scipy.stats.sampling import DiscreteAliasUrn
>>> class Distribution:
... def __init__(self, c):
... self.c = c
... def pmf(self, x):
... return x**self.c
... def support(self):
... return (0, 10)
...
>>> dist = Distribution(2)
>>> urng = np.random.default_rng()
>>> rng = DiscreteAliasUrn(dist, random_state=urng)
>>> rvs = rng.rvs(1000)
>>> fig = plt.figure()
>>> ax = fig.add_subplot(111)
>>> x = np.arange(1, 11)
>>> fx = dist.pmf(x)
>>> fx = fx / fx.sum()
>>> ax.plot(x, fx, 'bo', label='true distribution')
>>> ax.vlines(x, 0, fx, lw=2)
>>> ax.hist(rvs, bins=np.r_[x, 11]-0.5, density=True, alpha=0.5, color='r',
... label='samples')
>>> ax.set_xlabel('x')
>>> ax.set_ylabel('PMF(x)')
>>> ax.set_title('Discrete Alias Urn Samples')
>>> plt.legend()
>>> plt.show()
Note
As DiscreteAliasUrn expects PMF with signature
def pmf(self, x: float) -> float, it first vectorizes the
PMF using np.vectorize and then evaluates it over all the
points in the domain. But if the PMF is already vectorized,
it is much faster to just evaluate it at each point in the domain
and pass the obtained PV instead along with the domain.
For example, pmf methods of SciPy’s discrete distributions
are vectorized and a PV can be obtained by doing:
>>> from scipy.stats import binom
>>> from scipy.stats.sampling import DiscreteAliasUrn
>>> dist = binom(10, 0.2) # distribution object
>>> domain = dist.support() # the domain of your distribution
>>> x = np.arange(domain[0], domain[1] + 1)
>>> pv = dist.pmf(x)
>>> rng = DiscreteAliasUrn(pv, domain=domain)
Domain is required here to relocate the distribution.
The performance can be slightly influenced by setting the size of the used
table which can be changed by passing a urn_factor parameter.
>>> # use a table twice the length of PV.
>>> urn_factor = 2
>>> rng = DiscreteAliasUrn(pv, urn_factor=urn_factor, random_state=urng)
>>> rng.rvs()
2
Note
It is recommended to keep this parameter under 2.
Please see [1] and [2] for more details on this method.